$VAR1 = undef;
Summary for peptidase S01.174: mesotrypsin
| Names | |
|---|---|
| MEROPS Name | mesotrypsin |
| Other names | brain trypsin (Homo sapiens), mesotrypsinogen (activated), PRSS3 g.p. (Homo sapiens), PRSS4 g.p. (Homo sapiens), trypsin IV (Homo sapiens), trypsin-3 (Homo sapiens), trypsinogen-3 (Homo sapiens), trypsin-4 (Homo sapiens), trypsinogen IV, zymogen-X (activated) |
| Domain architecture |
|---|

| MEROPS Classification | |
|---|---|
| Classification | Clan PA >> Subclan PA(S) >> Family S1 >> Subfamily A >> S01.174 |
| Holotype | mesotrypsin (Homo sapiens), Uniprot accession P35030 (peptidase unit: 24-246), MERNUM MER0000022 |
| History | Identifier created: MEROPS 3.02 (25 June 1998) |
| Activity | |||
|---|---|---|---|
| Catalytic type | Serine | ||
| NC-IUBMB | Not yet included in IUBMB recommendations. | ||
| Proteolytic events | CutDB database (5 cleavages) | ||
| Pathways | KEGG | Influenza A | |
| KEGG | Neuroactive ligand-receptor interaction | ||
| KEGG | Pancreatic secretion | ||
| KEGG | Protein digestion and absorption | ||
| Other databases | TREEFAM | http://www.treefam.org/family/TF331065 | |
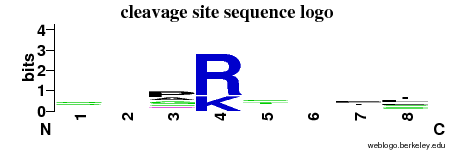
| Cleavage site specificity | Explanations of how to interpret the following cleavage site sequence logo and specificity matrix can be found here. | ||
| Cleavage pattern | -/-/pag/RK -/-/-/il (based on 22 cleavages) -/-/-/il (based on 22 cleavages) | ||
 |
| Specificity matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| Human genetics | ||||||
|---|---|---|---|---|---|---|
| Gene symbol | Locus | Megabases | Ensembl | Entrez gene | Gene Cards | OMIM |
| PRSS3 | 9p13 | ENSG00000010438 | 5646 | PRSS3 | ||
