$VAR1 = undef;
Summary for peptidase M14.002: carboxypeptidase A2
| Names | |
|---|---|
| MEROPS Name | carboxypeptidase A2 |
| Other names | CPA2 g.p. (Homo sapiens) |
| Domain architecture |
|---|

| MEROPS Classification | |
|---|---|
| Classification | Clan MC >> Subclan (none) >> Family M14 >> Subfamily A >> M14.002 |
| Holotype | carboxypeptidase A2 (Homo sapiens), Uniprot accession P48052 (peptidase unit: 115-419), MERNUM MER0001608 |
| History | Identifier created: Handbook of Proteolytic Enzymes (1998) Academic Press, London. |
| Activity | |||
|---|---|---|---|
| Catalytic type | Metallo | ||
| Peplist | Included in the Peplist with identifier PL00207 | ||
| NC-IUBMB | Subclass 3.4 (Peptidases) >> Sub-subclass 3.4.17 (Metallocarboxypeptidases) >> Peptidase 3.4.17.15 | ||
| Enzymology | BRENDA database | ||
| Activity status | human: active (Auld, 2004) mouse: active (by similarity) | ||
| Pathways | KEGG | Pancreatic secretion | |
| KEGG | Protein digestion and absorption | ||
| Other databases | WIKIPEDIA | http://en.wikipedia.org/wiki/Carboxypeptidase_A | |
| Cleavage site specificity | Explanations of how to interpret the following cleavage site sequence logo and specificity matrix can be found here. | ||
| Cleavage pattern | -/-/-/fg flw/-/-/- (based on 11 cleavages) flw/-/-/- (based on 11 cleavages) | ||
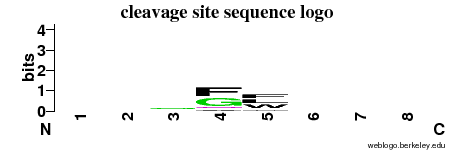 |
| Specificity matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| Human genetics | ||||||
|---|---|---|---|---|---|---|
| Gene symbol | Locus | Megabases | Ensembl | Entrez gene | Gene Cards | OMIM |
| CPA2 | 7q32 | ENSG00000158516 | 1358 | CPA2 | 600688 | |
| Mouse genetics | ||||||
| Gene symbol | Position | Megabases | Ensembl | Entrez gene | MGI | |
| Cpa2 | 6:A3.3 | ENSMUSG00000071553 | 232680 | MGI:3617840 | ||
