$VAR1 = undef;
Summary for peptidase A01.030: yapsin-1
| Names | |
|---|---|
| MEROPS Name | yapsin-1 |
| Other names | CgYps1 peptidase (Candida glabrata), YAP3 [obs.] g.p. (Saccharomyces cerevisiae), YPS1 g.p. (Saccharomyces cerevisiae), yeast aspartic protease 3 |
| Domain architecture |
|---|

| MEROPS Classification | |
|---|---|
| Classification | Clan AA >> Subclan (none) >> Family A1 >> Subfamily A >> A01.030 |
| Holotype | yapsin-1 (Saccharomyces cerevisiae), Uniprot accession P32329 (peptidase unit: 63-478), MERNUM MER0000948 |
| History | Identifier created: Handbook of Proteolytic Enzymes (1998) Academic Press, London. |
| Activity | |||
|---|---|---|---|
| Catalytic type | Aspartic | ||
| Peplist | Included in the Peplist with identifier PL00020 | ||
| NC-IUBMB | Subclass 3.4 (Peptidases) >> Sub-subclass 3.4.23 (Aspartic endopeptidases) >> Peptidase 3.4.23.41 | ||
| Enzymology | BRENDA database | ||
| Proteolytic events | CutDB database (4 cleavages) | ||
| Biotechnology | Can degrade proteins artificially expressed in Saccharomyces cerevisiae, so genetically-deficient strains may be advantageous (Kerry-Williams et al., 1998). | ||
| Physiology | Basic residue-specific processing of protein precursors in Saccharomyces cerevisiae (Ledgerwood et al., 1996). | ||
| Other databases | PANTHER | http://www.pantherdb.org/panther/familyList.do?searchType=basic&fieldName=all&listType=6&fieldValue=PTHR13683:SF412 | |
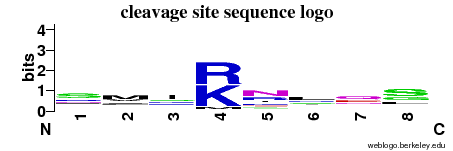
| Cleavage site specificity | Explanations of how to interpret the following cleavage site sequence logo and specificity matrix can be found here. | ||
| Cleavage pattern | s/m/i/RK nr/l/q/sgl (based on 46 cleavages) nr/l/q/sgl (based on 46 cleavages) | ||
 |
| Specificity matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
