$VAR1 = undef;
Summary for peptidase M02.006: angiotensin-converting enzyme-2
| Names | |
|---|---|
| MEROPS Name | angiotensin-converting enzyme-2 |
| Other names | ACE2, ACEH |
| Domain architecture |
|---|

| MEROPS Classification | |
|---|---|
| Classification | Clan MA >> Subclan MA(E) >> Family M2 >> Subfamily (none) >> M02.006 |
| Holotype | angiotensin-converting enzyme-2 (Homo sapiens), Uniprot accession Q9BYF1 (peptidase unit: 17-618), MERNUM MER0011061 |
| History | Identifier created: MEROPS 5.3 (4 December 2000) |
| Activity | |||
|---|---|---|---|
| Catalytic type | Metallo | ||
| Peplist | Included in the Peplist with identifier PL00135 | ||
| NC-IUBMB | Subclass 3.4 (Peptidases) >> Sub-subclass 3.4.17 (Metallocarboxypeptidases) >> Peptidase 3.4.17.23 | ||
| Enzymology | BRENDA database | ||
| Proteolytic events | CutDB database (3 cleavages) | ||
| Activity status | human: active (Turner & Hooper, 2004) mouse: active (by similarity) | ||
| Physiology | Genetic data show that this peptidase is an essential regulator of heart function at least in rodents and Drosophila (Crackower et al., 2002). Human angiotensin-converting enzyme 2 is a functional receptor for SARS coronavirus (Li et al., 2005). | ||
| Pathways | KEGG | Protein digestion and absorption | |
| KEGG | Renin-angiotensin system | ||
| Other databases | TREEFAM | http://www.treefam.org/family/TF312861 | |
| Cleavage site specificity | Explanations of how to interpret the following cleavage site sequence logo and specificity matrix can be found here. | ||
| Cleavage pattern | g/p/-/P frl/-/-/- (based on 14 cleavages) frl/-/-/- (based on 14 cleavages) | ||
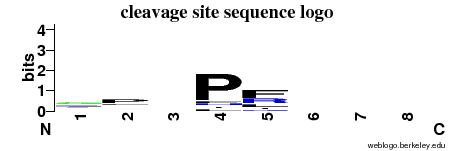 |
| Specificity matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| Human genetics | ||||||
|---|---|---|---|---|---|---|
| Gene symbol | Locus | Megabases | Ensembl | Entrez gene | Gene Cards | OMIM |
| ACE2 | Xp22 | ENSG00000130234 | 59272 | ACE2 | 300335 | |
| Mouse genetics | ||||||
| Gene symbol | Position | Megabases | Ensembl | Entrez gene | MGI | |
| Ace2 | X:F5 | ENSMUSG00000015405 | 70008 | MGI:1917258 | ||
