$VAR1 = undef;
Summary for peptidase M16.003: mitochondrial processing peptidase beta-subunit
| Names | |
|---|---|
| MEROPS Name | mitochondrial processing peptidase beta-subunit |
| Other names | MPP, PMPCB g.p. (Homo sapiens), processing enhancing protein |
| Domain architecture |
|---|

| MEROPS Classification | |
|---|---|
| Classification | Clan ME >> Subclan (none) >> Family M16 >> Subfamily B >> M16.003 |
| Holotype | mitochondrial processing peptidase beta-subunit (Saccharomyces cerevisiae), Uniprot accession P10507 (peptidase unit: 30-245), MERNUM MER0001229 |
| History | Identifier created: Handbook of Proteolytic Enzymes (1998) Academic Press, London. |
| Activity | |||
|---|---|---|---|
| Catalytic type | Metallo | ||
| Peplist | Included in the Peplist with identifier PL00222 | ||
| NC-IUBMB | Subclass 3.4 (Peptidases) >> Sub-subclass 3.4.24 (Metalloendopeptidases) >> Peptidase 3.4.24.64 | ||
| Enzymology | BRENDA database | ||
| Physiology | Processes N-terminal targetting peptides from mitochondrial proteins during import. | ||
| Other databases | TREEFAM | http://www.treefam.org/family/TF105032 | |
| Cleavage site specificity | Explanations of how to interpret the following cleavage site sequence logo and specificity matrix can be found here. | ||
| Cleavage pattern | -/r/R/- ylfa/sa/-/- (based on 86 cleavages) ylfa/sa/-/- (based on 86 cleavages) | ||
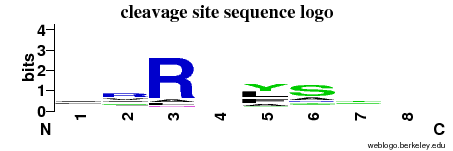 |
| Specificity matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
