$VAR1 = undef;
Summary for peptidase C14.004: caspase-7
| Names | |
|---|---|
| MEROPS Name | caspase-7 |
| Other names | caspase 7, ICE-LAP3, Mch3, CMH-1 |
| Domain architecture |
|---|

| MEROPS Classification | |
|---|---|
| Classification | Clan CD >> Subclan (none) >> Family C14 >> Subfamily A >> C14.004 |
| Holotype | caspase-7 (Homo sapiens), Uniprot accession P55210 (peptidase unit: 62-336), MERNUM MER0002705 |
| History | Identifier created: MEROPS 3.02 (25 June 1998) |
| Activity | |||
|---|---|---|---|
| Catalytic type | Cysteine | ||
| Peplist | Included in the Peplist with identifier PL00101 | ||
| NC-IUBMB | Subclass 3.4 (Peptidases) >> Sub-subclass 3.4.22 (Cysteine endopeptidases) >> Peptidase 3.4.22.60 | ||
| Enzymology | BRENDA database | ||
| Proteolytic events | CutDB database (53 cleavages) | ||
| Activity status | human: active (Nicholson & Thornberry, 2004) mouse: active (by similarity) (Lee et al., 2001) | ||
| Physiology | One of the "executioner" caspases in mammalian cell apoptosis. | ||
| Pharmaceutical relevance | A non-peptidic inhibitor of caspase-3 and caspase-7 was effective in reducing cardiac ischemic injury in a rabbit model (Chapman et al., 2002). | ||
| Pathways | KEGG | Alzheimer's disease | |
| KEGG | Apoptosis | ||
| KEGG | Legionellosis | ||
| KEGG | Non-alcoholic fatty liver disease (NAFLD) | ||
| KEGG | Pertussis | ||
| KEGG | TNF signaling pathway | ||
| Other databases | WIKIPEDIA | http://en.wikipedia.org/wiki/Caspase_7 | |
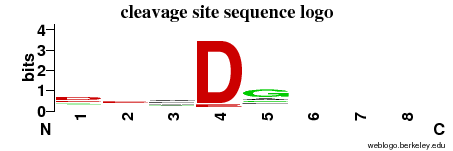
| Cleavage site specificity | Explanations of how to interpret the following cleavage site sequence logo and specificity matrix can be found here. | ||
| Cleavage pattern | d/-/-/D g/-/-/- (based on 502 cleavages) g/-/-/- (based on 502 cleavages) | ||
 |
| Specificity matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| Specificity from combinatorial peptides | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| Human genetics | ||||||
|---|---|---|---|---|---|---|
| Gene symbol | Locus | Megabases | Ensembl | Entrez gene | Gene Cards | OMIM |
| CASP7 | 10q25.1-q25.2 | ENSG00000165806 | 840 | CASP7 | 601761 | |
| Mouse genetics | ||||||
| Gene symbol | Position | Megabases | Ensembl | Entrez gene | MGI | |
| Casp7 | 19:D2 | ENSMUSG00000025076 | 12369 | MGI:109383 | ||
 Gxxx
Gxxx