| Activity |
|---|
| Catalytic type | Metallo |
| Peplist | Included in the Peplist with identifier PL00171 |
| NC-IUBMB | Subclass 3.4 (Peptidases) >> Sub-subclass 3.4.24 (Metalloendopeptidases) >> Peptidase 3.4.24.40
|
| Enzymology | BRENDA database |
| Physiology | Enhances attachment of organism to corneal epithelium, playing a major role in pathogenesis. |
| Pharmaceutical relevance | Target for vaccine development, and chemotherapy of bacterial infection. |
|
Other databases
| PANTHER | http://www.pantherdb.org/panther/familyList.do?searchType=basic&fieldName=all&listType=6&fieldValue=PTHR36921:SF3 |
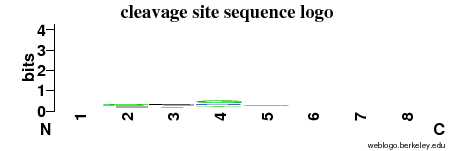
| Cleavage site specificity |
Explanations of how to interpret the
following cleavage site sequence logo and specificity matrix can be found here. |
|---|
| Cleavage pattern | -/-/-/g -/-/-/- (based on 13 cleavages) -/-/-/- (based on 13 cleavages) |

 -/-/-/- (based on 13 cleavages)
-/-/-/- (based on 13 cleavages)