$VAR1 = undef;
Summary for peptidase C13.009: butelase 1
| Names | |
|---|---|
| MEROPS Name | butelase 1 |
| Other names | butelase 2 |
| Domain architecture |
|---|

| MEROPS Classification | |
|---|---|
| Classification | Clan CD >> Subclan (none) >> Family C13 >> Subfamily (none) >> C13.009 |
| Holotype | butelase 1 (Clitoria ternatea) (peptidase unit: 42-318), MERNUM MER0511082 |
| History | Identifier created: MEROPS 11.0 (6 January 2017) |
| Activity | |||
|---|---|---|---|
| Catalytic type | Cysteine | ||
| NC-IUBMB | Not yet included in IUBMB recommendations. | ||
| Cleavage site specificity | Explanations of how to interpret the following cleavage site sequence logo and specificity matrix can be found here. | ||
| Cleavage pattern | Cs/yc/krn/N H/V/Ic/Ay (based on 12 cleavages) H/V/Ic/Ay (based on 12 cleavages) | ||
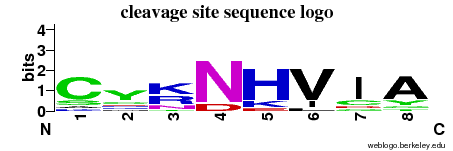 |
| Specificity matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
