$VAR1 = undef;
Summary for peptidase S10.009: serine carboxypeptidase III (plant)
| Names | |
|---|---|
| MEROPS Name | serine carboxypeptidase III (plant) |
| Other names | wheat carboxypeptidase III |
| Domain architecture |
|---|

| MEROPS Classification | |
|---|---|
| Classification | Clan SC >> Subclan (none) >> Family S10 >> Subfamily (none) >> S10.009 |
| Holotype | serine carboxypeptidase III (plant) (Hordeum vulgare), Uniprot accession P21529 (peptidase unit: 82-492), MERNUM MER0001760 |
| History | Identifier created: MEROPS 5.61 (29 October 2001) |
| Activity | |||
|---|---|---|---|
| Catalytic type | Serine | ||
| NC-IUBMB | Not yet included in IUBMB recommendations. | ||
| Other databases | PANTHER | http://www.pantherdb.org/panther/familyList.do?searchType=basic&fieldName=all&listType=6&fieldValue=PTHR11802:SF191 | |
| Cleavage site specificity | Explanations of how to interpret the following cleavage site sequence logo and specificity matrix can be found here. | ||
| Cleavage pattern | -/-/-/A -/-/-/- (based on 16 cleavages) -/-/-/- (based on 16 cleavages) | ||
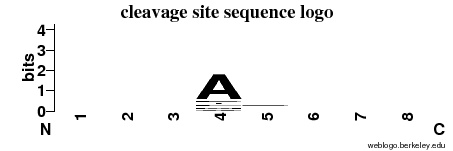 |
| Specificity matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
