Do data resources managed by EMBL-EBI and our collaborators make a difference to your work? Please take 10 minutes to fill in our annual user survey, and help us make the case for why sustaining open data resources is critical for life sciences research. We appreciate your responses prior to closing of the survey on Monday 17th June.
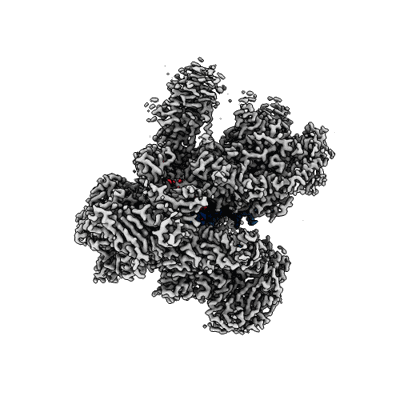
Structure of yeast RNA Polymerase III Pre-Termination Complex (PTC)
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 7z1l
Q-score: 0.538
Girbig M, Xie J, Grotsch H, Libri D, Porrua O, Muller CW
Cell Rep (2022) 40 pp. 111316-111316 [ Pubmed: 36070694 DOI: doi:10.1016/j.celrep.2022.111316 ]
- Dna-directed rna polymerases i and iii subunit rpac2 (16 kDa, Protein from Saccharomyces cerevisiae W303)
- (3r,5s,7r,8r,9s,10s,12s,13r,14s,17r)-10,13-dimethyl-17-[(2r)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1h-cyclopenta[a]phenanthrene-3,7,12-triol (378 Da, Ligand)
- T-dna (13 kDa, DNA from synthetic construct)
- Dna-directed rna polymerase iii subunit rpc5 (32 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc3 (74 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc2 (17 kDa, Protein from Saccharomyces cerevisiae W303)
- Zinc ion (65 Da, Ligand)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc4 (7 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc1 (162 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc10 (12 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc1 (25 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc7 (27 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i and iii subunit rpac1 (37 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc2 (129 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc3 (16 kDa, Protein from Saccharomyces cerevisiae W303)
- Nt-dna (13 kDa, DNA from synthetic construct)
- Dna-directed rna polymerase iii subunit rpc8 (24 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc4 (46 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc6 (36 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc5 (8 kDa, Protein from Saccharomyces cerevisiae W303)
- Yeast rna polymerase iii ptc (730 kDa, Complex from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc9 (18 kDa, Protein from Saccharomyces cerevisiae W303)
- Rna (7 kDa, RNA from synthetic construct)
- Magnesium ion (24 Da, Ligand)
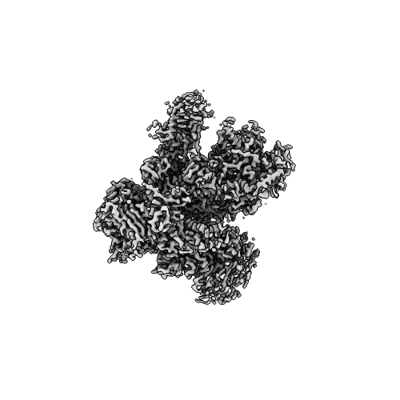
Structure of yeast RNA Polymerase III Elongation Complex (EC)
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7z1m
Q-score: 0.466
Girbig M, Xie J, Grotsch H, Libri D, Porrua O, Muller CW
Cell Rep (2022) 40 pp. 111316-111316 [ Pubmed: 36070694 DOI: doi:10.1016/j.celrep.2022.111316 ]
- Dna-directed rna polymerases i and iii subunit rpac2 (16 kDa, Protein from Saccharomyces cerevisiae W303)
- (3r,5s,7r,8r,9s,10s,12s,13r,14s,17r)-10,13-dimethyl-17-[(2r)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1h-cyclopenta[a]phenanthrene-3,7,12-triol (378 Da, Ligand)
- Dna-directed rna polymerase iii subunit rpc5 (32 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc3 (74 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc2 (17 kDa, Protein from Saccharomyces cerevisiae W303)
- Zinc ion (65 Da, Ligand)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc4 (7 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc1 (162 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc10 (12 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc1 (25 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc7 (27 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i and iii subunit rpac1 (37 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc2 (129 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc3 (16 kDa, Protein from Saccharomyces cerevisiae W303)
- T-dna (13 kDa, Macromolecule from synthetic construct)
- Dna-directed rna polymerase iii subunit rpc8 (24 kDa, Protein from Saccharomyces cerevisiae W303)
- Yeast rna polymerase iii ec (730 kDa, Complex from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc4 (46 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc6 (36 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc5 (8 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc9 (18 kDa, Protein from Saccharomyces cerevisiae W303)
- Nt-dna (13 kDa, Macromolecule from synthetic construct)
- Rna (6 kDa, RNA from synthetic construct)
- Magnesium ion (24 Da, Ligand)
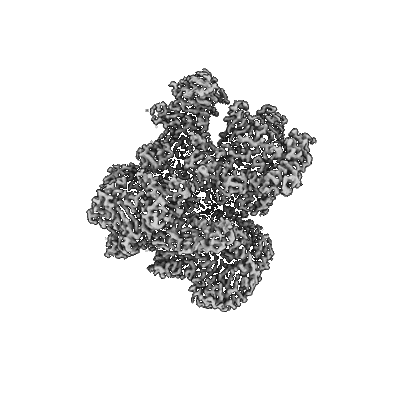
Structure of yeast RNA Polymerase III Delta C53-C37-C11
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 7z1n
Q-score: 0.405
Girbig M, Xie J, Grotsch H, Libri D, Porrua O, Muller CW
Cell Rep (2022) 40 pp. 111316-111316 [ Pubmed: 36070694 DOI: doi:10.1016/j.celrep.2022.111316 ]
- Dna-directed rna polymerases i and iii subunit rpac2 (16 kDa, Protein from Saccharomyces cerevisiae W303)
- T-dna (13 kDa, DNA from synthetic construct)
- Dna-directed rna polymerase iii subunit rpc3 (74 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc2 (17 kDa, Protein from Saccharomyces cerevisiae W303)
- Zinc ion (65 Da, Ligand)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc4 (7 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc1 (162 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc1 (25 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc7 (27 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i and iii subunit rpac1 (37 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc2 (129 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc3 (16 kDa, Protein from Saccharomyces cerevisiae W303)
- Nt-dna (13 kDa, DNA from synthetic construct)
- Dna-directed rna polymerase iii subunit rpc8 (24 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc6 (36 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc5 (8 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc9 (18 kDa, Protein from Saccharomyces cerevisiae W303)
- Rna (7 kDa, RNA from synthetic construct)
- Yeast rna polymerase iii delta c53-c37-c11 (635 kDa, Complex from Saccharomyces cerevisiae W303)
- Magnesium ion (24 Da, Ligand)
- Chapso (631 Da, Ligand)
Structure of yeast RNA Polymerase III PTC + NTPs
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 7z1o
Q-score: 0.534
Girbig M, Xie J, Grotsch H, Libri D, Porrua O, Muller CW
Cell Rep (2022) 40 pp. 111316-111316 [ Pubmed: 36070694 DOI: doi:10.1016/j.celrep.2022.111316 ]
- Dna-directed rna polymerases i and iii subunit rpac2 (16 kDa, Protein from Saccharomyces cerevisiae W303)
- T-dna (13 kDa, DNA from synthetic construct)
- Dna-directed rna polymerase iii subunit rpc5 (32 kDa, Protein from Saccharomyces cerevisiae W303)
- Yeast rna polymerase iii ptc + ntps (730 kDa, Complex from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc2 (17 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc3 (74 kDa, Protein from Saccharomyces cerevisiae W303)
- Zinc ion (65 Da, Ligand)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc4 (7 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc1 (162 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc10 (12 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc1 (25 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc7 (27 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i and iii subunit rpac1 (37 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc2 (129 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc3 (16 kDa, Protein from Saccharomyces cerevisiae W303)
- Nt-dna (13 kDa, DNA from synthetic construct)
- Dna-directed rna polymerase iii subunit rpc8 (24 kDa, Protein from Saccharomyces cerevisiae W303)
- Rna (6 kDa, RNA from synthetic construct)
- Dna-directed rna polymerase iii subunit rpc4 (46 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc6 (36 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc5 (8 kDa, Protein from Saccharomyces cerevisiae W303)
- Dna-directed rna polymerase iii subunit rpc9 (18 kDa, Protein from Saccharomyces cerevisiae W303)
- Magnesium ion (24 Da, Ligand)
- Chapso (631 Da, Ligand)
Cryo-EM structure of human apo RNA Polymerase III
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 7a6h
Q-score: 0.425
Girbig M, Misiaszek AD, Vorlander MK, Lafita A, Grotsch H, Baudin F, Bateman A, Muller CW
Nat Struct Mol Biol (2021) 28 pp. 210-219 [ DOI: doi:10.1038/s41594-020-00555-5 Pubmed: 33558764 ]
- Dna-directed rna polymerases i and iii subunit rpac2 (15 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc4 (7 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc9 (16 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc10 (12 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc2 (14 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc1 (24 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc5 (80 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc2 (127 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc3 (17 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc3 (60 kDa, Protein from Homo sapiens)
- Iron/sulfur cluster (351 Da, Ligand)
- Dna-directed rna polymerases i and iii subunit rpac1 (39 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc4 (44 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc5 (7 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc8 (22 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc7 (25 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc6 (35 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Cryo-em structure of human apo rna polymerase iii (776 kDa, Complex from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc1 (155 kDa, Protein from Homo sapiens)
Cryo-EM structure of human RNA Polymerase III elongation complex 1
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 7ae1
Q-score: 0.48
Girbig M, Misiaszek AD, Vorlander MK, Lafita A, Grotsch H, Baudin F, Bateman A, Muller CW
Nat Struct Mol Biol (2021) 28 pp. 210-219 [ DOI: doi:10.1038/s41594-020-00555-5 Pubmed: 33558764 ]
- Dna-directed rna polymerases i and iii subunit rpac2 (15 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc4 (7 kDa, Protein from Homo sapiens)
- Template-dna (14 kDa, DNA from synthetic construct)
- Dna-directed rna polymerase iii subunit rpc9 (16 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc10 (12 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc2 (14 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc1 (24 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc5 (80 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc2 (127 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc3 (17 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc3 (60 kDa, Protein from Homo sapiens)
- Rna, dna (Complex from synthetic construct)
- Iron/sulfur cluster (351 Da, Ligand)
- Dna-directed rna polymerases i and iii subunit rpac1 (39 kDa, Protein from Homo sapiens)
- Human rna polymerase iii elongation complex 1 (776 kDa, Complex)
- Human rna polymerase (Complex from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc4 (44 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc5 (7 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc8 (22 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc7 (25 kDa, Protein from Homo sapiens)
- Non-template dna (14 kDa, DNA from synthetic construct)
- Dna-directed rna polymerase iii subunit rpc6 (35 kDa, Protein from Homo sapiens)
- Rna (6 kDa, RNA from synthetic construct)
- Magnesium ion (24 Da, Ligand)
- Dna-directed rna polymerase iii subunit rpc1 (155 kDa, Protein from Homo sapiens)
Cryo-EM structure of human RNA Polymerase III elongation complex 3
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 7ae3
Q-score: 0.373
Girbig M, Misiaszek AD, Vorlander MK, Lafita A, Grotsch H, Baudin F, Bateman A, Muller CW
Nat Struct Mol Biol (2021) 28 pp. 210-219 [ DOI: doi:10.1038/s41594-020-00555-5 Pubmed: 33558764 ]
- Dna-directed rna polymerases i and iii subunit rpac2 (15 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc4 (7 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc9 (16 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc10 (12 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc2 (14 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc1 (24 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc5 (80 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc2 (127 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc3 (17 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc3 (60 kDa, Protein from Homo sapiens)
- Human rna polymerase iii elongation complex 3 (776 kDa, Complex)
- Rna, dna (Complex from synthetic construct)
- Template dna (14 kDa, DNA from synthetic construct)
- Iron/sulfur cluster (351 Da, Ligand)
- Human rna polymerase iii elongation complex (Complex from Homo sapiens)
- Dna-directed rna polymerases i and iii subunit rpac1 (39 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc4 (44 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc5 (7 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc8 (22 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc7 (25 kDa, Protein from Homo sapiens)
- Non-template dna (14 kDa, DNA from synthetic construct)
- Dna-directed rna polymerase iii subunit rpc6 (35 kDa, Protein from Homo sapiens)
- Rna (6 kDa, RNA from synthetic construct)
- Magnesium ion (24 Da, Ligand)
- Dna-directed rna polymerase iii subunit rpc1 (155 kDa, Protein from Homo sapiens)
Cryo-EM structure of human RNA Polymerase III elongation complex 2
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7aea
Q-score: 0.409
Girbig M, Misiaszek AD, Vorlander MK, Lafita A, Grotsch H, Baudin F, Bateman A, Muller CW
Nat Struct Mol Biol (2021) 28 pp. 210-219 [ DOI: doi:10.1038/s41594-020-00555-5 Pubmed: 33558764 ]
- Dna-directed rna polymerases i and iii subunit rpac2 (15 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc4 (7 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc9 (16 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc10 (12 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc2 (14 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc1 (24 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc5 (80 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc2 (127 kDa, Protein from Homo sapiens)
- Human rna polymerase iii elongation complex (776 kDa, Complex)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc3 (17 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc3 (60 kDa, Protein from Homo sapiens)
- Rna, dna (Complex from synthetic construct)
- Template dna (14 kDa, DNA from synthetic construct)
- Iron/sulfur cluster (351 Da, Ligand)
- Human rna polymerase iii elongation complex (Complex from Homo sapiens)
- Dna-directed rna polymerases i and iii subunit rpac1 (39 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc4 (44 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerases i, ii, and iii subunit rpabc5 (7 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc8 (22 kDa, Protein from Homo sapiens)
- Dna-directed rna polymerase iii subunit rpc7 (25 kDa, Protein from Homo sapiens)
- Non-template dna (14 kDa, DNA from synthetic construct)
- Dna-directed rna polymerase iii subunit rpc6 (35 kDa, Protein from Homo sapiens)
- Rna (6 kDa, RNA from synthetic construct)
- Magnesium ion (24 Da, Ligand)
- Dna-directed rna polymerase iii subunit rpc1 (155 kDa, Protein from Homo sapiens)