 FROG analysis - a community standard to foster reproducibility and curation of constraint-based models
FROG analysis - a community standard to foster reproducibility and curation of constraint-based models
Summary
Constraint-based models are used to investigate metabolism in diverse conditions; particularly genome-scale metabolic models (GEMs) with thousands of reactions provide opportunities to analyze organism-specific metabolism. Community standards for consistent model reconstruction, curation, and sharing are crucial to ensure reproducibility, reliability, and FAIR sharing of the models. MEMOTE, a community tool was developed for standardized quality assessment of the models. Currently, it is not possible to assess whether constraint-based models including Genome-Scale metabolic models (GEM) are reproducible because these models often have multiple solutions and the numerical values are not always enumerated in the published manuscripts. Here we initiate a community effort for standardized assessment of model reproducibility, which is currently lacking. Reproducibility of results is the cornerstone of science and its assessment is an essential part of the curation of a model in the repository such as BioModels. Following discussions at dedicated breakout sessions at HARMONY2020 (minutes of the meeting), COMBINE2020 (minutes of the meeting), and HARMONY2021 (minutes of the meeting) we have now developed FROG analysis, an ensemble of analysis of constraint-based models to generate standardized numerically reproducible reference dataset (aka FROG report). We have also developed a collection of tools that generates FROG reports in a standardized schema. We propose that the modelers share FROG reports along with their model which can be used by the modeler or curator to independently assess the reproducibility of a model. FROG analysis is currently used in BioModels’ workflow for the curation of constraint-based models along with the MEMOTE test suite. To allow retrospective curation of previously published models, we propose that the model authors submit a miniFROG report in addition to autogenerated FROG report. The miniFROG report is a manually created data table in a standardized schema that lists at least a couple of results described in the manuscript and compares them against the model results from the FROG report. Sharing FROG and miniFROG reports along with model (SBML-FBC) files will enable assessment of reproducibility and curation of models and thereby greatly enhance reuse, extension, integration of constraint-based models for new knowledge generation.
FROG analysis

1) Objective Function Values
The objective function value for a defined set of bounds should be comparable/reproducible.
Some models may not have objective function values, in such cases, a list of common objective functions can be used. Couple of examples are listed below
Objective functions of internal fluxes in E.coli
Schuetz et al. (2007) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1949037/
- A detailed discussion on the biomass objectives
Feist and Palsson (2010) https://www.sciencedirect.com/science/article/abs/pii/S1369527410000512
2) Flux Variability Analysis (FVA)
- FVA span: min/max of flux should be comparable (for a particular objective function value)
- These values will have only small numerical differences among software, depending on the set boundary conditions.
3) Gene Deletion Fluxes
- The systematic deletion of all genes one at a time should provide comparable reference results.
4) Reaction Deletion (extended coverage of reaction network)
- The systematic deletion of all reactions one at a time should provide comparable reference results.
The above FROG references will be central to assessing the reproducibility of the model and the curation efforts.
FROG test suite
The following tools can be used to generate a FROG report.
- FROG Report Generator (Devekioers: Mirek Kratochvíl and St. Elmo Wilken)
FBCModelTests.jl a Julia package based on COBREXA
Command-line Tool: https://github.com/LCSB-BioCore/FBCModelTests.jl
Web implementation: https://fbc-model-tests.lcsb.uni.lu/
- FBC Curation Matlab Toolbox (Developer: Karthik Raman)
MATLAB/COBRA helper for FROG analysis of FBC models.
Command-line Tool: https://github.com/RamanLab/fbc_curation_matlab
- runFROG (Developer: Matthias König) (tool not updated)
a python package for FROG analysis. Currently it includes two separate implementations of the reference files generation by:
(a)cobrapy based on COBRApy (Constraint-Based Reconstruction and Analysis in Python) available from https://github.com/opencobra/cobrapy/
(b) cameo based on Cameo (Cameo—Computer Aided Metabolic Engineering and Optimization) available from https://github.com/biosustain/cameo
Command-line Tool: https://github.com/matthiaskoenig/fbc_curation
Documentation: https://fbc-curation.readthedocs.io/en/latest/index.html
Web implementation: http://runfrog.de/
- CBMPy model curator (Developer: Brett Oliver)
Script and web-based implementation of the FROG analysis using CBMPy.
Web implementation: CBMPyWEB
Command-line Tool: CBMPy FBC curator https://github.com/matthiaskoenig/fbc_curation
Documentation: link to presentation
- FLUXER (tool not updated)
a web-based implementation supporting FROG analysis using the runFROG python package in the backend.
Web page: https://fluxer.umbc.edu/
Curation of FBC models in BioModels
BioModels is one of the largest repositories of manually curated models. Model curation involves ensuring the model is (1) encoded in a syntactically valid standard format such as SBML, (2) is and reproducible, and (3) semantically enriched with controlled vocabularies such GO, ChEBI, etc. A model author is expected to submit an SBML model (as main file), FROG report (as additional file), and miniFROG (as additional file) to BioModels. Reproducibility of the constraint-based SBML models submitted to BioModels will be verified using the FROG test suite. Curators at BioModels will independently try to reproduce the FROG report using a tool different from the one used by the modeler. If the results are reproducible, the model will be added to the curated branch of BioModels. The quality of the model will be tested using MEMOTE test suite and the report will be uploaded as an additional file. Furthermore, model-level semantic annotations will be added to the model following MIRIAM guidelines. Refer to Figure 1 for the complete workflow.
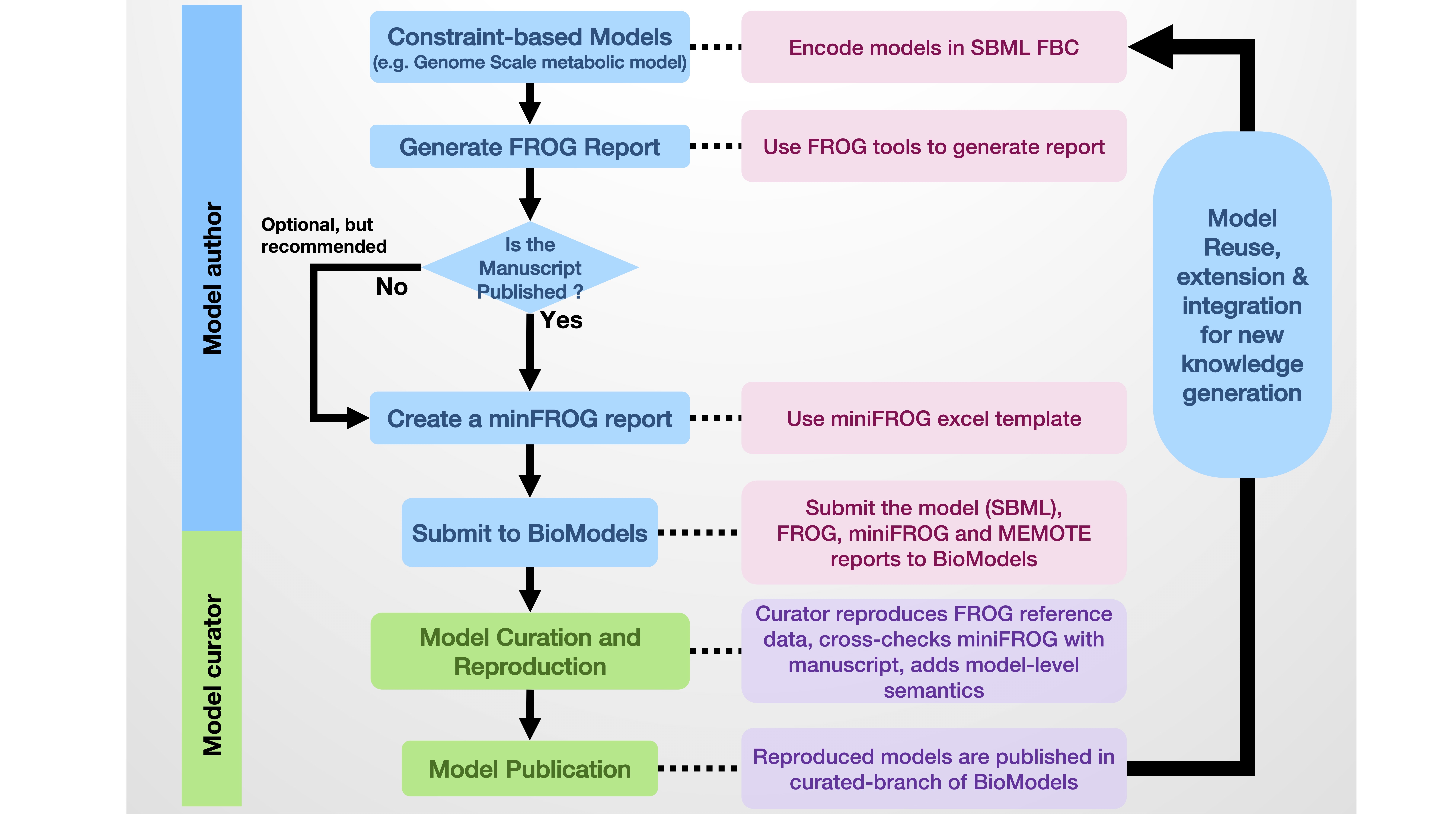
Figure 1: Workflow for the curation of constraint-based models in BioModels using FROG
minFROG description

Following are the fields in the miniFROG:
Publication Information
Organism Name
BioModel Identifier
Simulation constraints defined in publication
Gene/ reaction involved in the constraint
Type of constraints in the simulation
Tools used for simulation
Results from publication
Results predicted from FROG
Type of F/R/O/G analysis
Line in FROG Report
Validation of the simulation (Yes/No)
Remarks
miniFROG report should be manually prepared using this template and submitted as an additional file during model submission to BioModels for all previously published models. miniFROG report is optional, but highly recommended and can be submitted as an additional file for unpublished models.
Raman et al. 2005 model (BIOMD0000001046) is curated using FROG test suite and miniFROG standards. It provides an example for miniFROG report. Click here to view other examples on miniFROG reports.
Community manuscript
We are preparing a manuscript “FROG analysis - a community standard to foster reproducibility and curation of constraint-based models”
If you like to contribute to this community effort and the manuscript,
Please submit one of your constraint-based models (to BioModels*) along with a FROG report generated using any one of the above FROG tools or your own tool in the standard schema.
Contact: biomodels-cura@ebi.ac.uk for any support on submission. While contacting, use the keyword 'FROG' in the subject line.
* If you have already submitted your model to BioModels, kindly upload the FROG report as an additional file and request to publish the model again.
Please fill the following form below
https://docs.google.com/forms/d/e/1FAIpQLSca8t77y_bn85zEbVvKBtylN_UnHSurKBCnPTtMTpyVBnpBZA/viewform
All models submitted to BioModels will be curated using the FROG tools, added to the appropriate branch and highlighted in the manuscript.
Develop a package or web application for FROG analysis.
Please contact: sheriff@ebi.ac.uk
