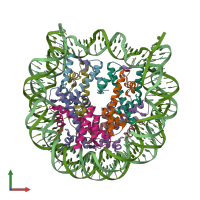
Assemblies
Assembly Name:
Nucleosome, variant H3.1-H2A.V-H2B.1 and DNA
Multimeric state:
hetero decamer
Accessible surface area:
72576.53 Å2
Buried surface area:
55912.57 Å2
Dissociation area:
5,804.68
Å2
Dissociation energy (ΔGdiss):
93.8
kcal/mol
Dissociation entropy (TΔSdiss):
16.11
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-117076
Macromolecules
Chains: A, E
Length: 139 amino acids
Theoretical weight: 15.72 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: Core histone H2A/H2B/H3/H4
InterPro:
CATH: Histone, subunit A
Length: 139 amino acids
Theoretical weight: 15.72 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 P68431 (Residues: 1-136; Coverage: 100%)
P68431 (Residues: 1-136; Coverage: 100%)
Pfam: Core histone H2A/H2B/H3/H4
InterPro:
CATH: Histone, subunit A
Chains: B, F
Length: 106 amino acids
Theoretical weight: 11.68 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: Centromere kinetochore component CENP-T histone fold
InterPro:
Length: 106 amino acids
Theoretical weight: 11.68 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 P62805 (Residues: 1-103; Coverage: 100%)
P62805 (Residues: 1-103; Coverage: 100%)
Pfam: Centromere kinetochore component CENP-T histone fold
InterPro:
- Histone-fold
- Histone H4
- Histone H4, conserved site
- TATA box binding protein associated factor (TAF), histone-like fold domain
- CENP-T/Histone H4, histone fold
Chains: C, G
Length: 131 amino acids
Theoretical weight: 13.82 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam:
InterPro:
Length: 131 amino acids
Theoretical weight: 13.82 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q71UI9 (Residues: 1-128; Coverage: 100%)
Q71UI9 (Residues: 1-128; Coverage: 100%)
Pfam:
InterPro:
- Histone H2A/H2B/H3
- Histone-fold
- Histone H2A
- Histone H2A conserved site
- Histone H2A, C-terminal domain
Chains: D, H
Length: 129 amino acids
Theoretical weight: 14.22 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: Core histone H2A/H2B/H3/H4
InterPro:
CATH: Histone, subunit A
Length: 129 amino acids
Theoretical weight: 14.22 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 P06899 (Residues: 1-126; Coverage: 100%)
P06899 (Residues: 1-126; Coverage: 100%)
Pfam: Core histone H2A/H2B/H3/H4
InterPro:
CATH: Histone, subunit A
Name:
DNA (146-MER)
Representative chains: I, J
Length: 146 nucleotides
Theoretical weight: 45.05 KDa
Representative chains: I, J
Length: 146 nucleotides
Theoretical weight: 45.05 KDa