- Course overview
- Search within this course
- Introduction
- Real-time PCR
- What is Next Generation DNA Sequencing?
- RNA sequencing
- Biological interpretation of gene expression data
- Genotyping, epigenetic and DNA/RNA-protein interaction methods
- DNA/RNA-protein interactions
- Summary
- Quiz: Check your learning
- Your feedback
- Learn more
- References
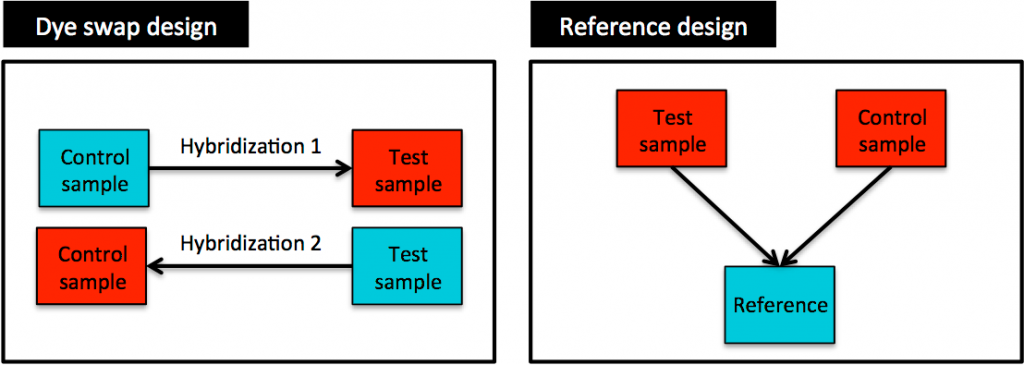
Counteracting dye bias effects when using two-colour arrays
One issue for two-colour arrays is related to dye bias effects introduced by the slightly different chemistry of the two dyes. It is important to control for this dye bias in the design of your experiment, for example by using a dye swap design or a reference design (Figure 3). In a dye swap design, the same pairs of samples (test and control) are compared twice with the dye assignment reversed in the second hybridisation. However, the most common design for two colour microarrays is the reference design in which each experimental sample is hybridised against a common reference sample.
Performing replicates
Replicates are essential for reliably detecting differentially expressed genes in microarray experiments. Without replicates, no statistical analysis of the significance and reliability of the observed changes is possible; the typical result is an increased number of both false-positive and false-negative errors in detecting differentially expressed genes. There are two types of replicates: technical replicates and biological replicates (see the section on Number of replicates later in the course).
In Expression Atlas, a minimum acceptable number of biological sample replicates (three) is enforced to ensure sufficient statistical power to detect differential expression (3).