Epigenetic modifications
Epigenetic modifications are heritable chemical or physical changes in chromatin. There are two types of epigenetic modifications – DNA methylation and histone modifications (16).
DNA methylation
DNA methylation involves the addition of a methyl or hydroxymethyl group to bases in the DNA sequence. The most commonly studied modification is the methylation of the C5 position on cytosine bases, or m5c (17,18).
DNA methylation can be measured using chromatin immunoprecipitation (ChIP) or bisulfite-based methods (Figure 14).
ChIP-based methods
ChIP-based methods use methylation-specific antibodies to purify methylated regions of the genome. The DNA is then analysed by microarray or NGS to identify these regions (17,18).
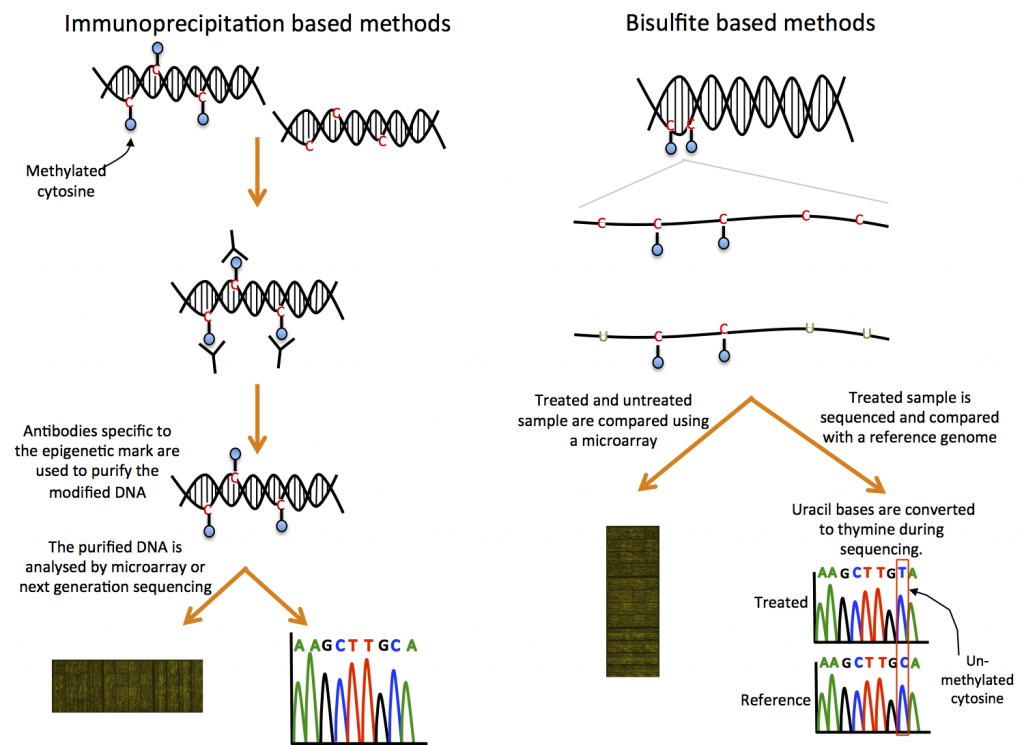
Bisulphite-based methods
Bisulfite-based methods involve bisulfite treatment of the DNA sample, which converts unmethylated cytosine bases to a uracil base, while leaving methylated residues as a cytosine. The treated DNA is then analysed by microarray or NGS. This method can only detect types of methylation that are susceptible to bisulfite-induced changes (17,18).
For microarray analysis, the bisulfite-treated sample is competitively hybridised with an untreated sample. The signal intensity ratio between the samples is used to infer the level of methylation in specific regions of the genome.
For NGS, the treated and untreated samples are sequenced and compared to identify specific methylation sites within the genome. Bisulfite sequencing requires a well annotated genome.
Epigenetic modifications of RNA such as m6A methylation have recently been described and are thought to regulate RNA stability and localisation (19). These modifications can not be detected by bisulfite-based methods. They can be detected by MeRIP-Seq (methylated RNA immunoprecipitation sequencing), which combines immunoprecipitation of modified sequences with RNA-seq analysis.