 |
PDBsum entry 1xff

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.6.1.16
- glutamine--fructose-6-phosphate transaminase (isomerizing).
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
UDP-N-acetylglucosamine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-fructose 6-phosphate + L-glutamine = D-glucosamine 6-phosphate + L-glutamate
|
 |
 |
 |
 |
 |
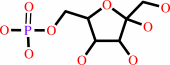 D-fructose 6-phosphate
D-fructose 6-phosphate
|
+
|
 L-glutamine
L-glutamine
|
=
|
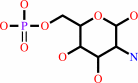 D-glucosamine 6-phosphate
D-glucosamine 6-phosphate
|
+
|
L-glutamate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
4:801-810
(1996)
|
|
PubMed id:
|
|
|
|

|
| |
|
Substrate binding is required for assembly of the active conformation of the catalytic site in Ntn amidotransferases: evidence from the 1.8 A crystal structure of the glutaminase domain of glucosamine 6-phosphate synthase.
|
|
M.N.Isupov,
G.Obmolova,
S.Butterworth,
M.A.Badet-Denisot,
B.Badet,
I.Polikarpov,
J.A.Littlechild,
A.Teplyakov.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: Amidotransferases use the amide nitrogen of glutamine in a number of
important biosynthetic reactions. They are composed of a glutaminase domain,
which catalyzes the hydrolysis of glutamine to glutamate and ammonia, and a
synthetase domain, catalyzing amination of the substrate. To gain insight into
the mechanism of nitrogen transfer, we examined the structure of the glutaminase
domain of glucosamine 6-phosphate synthase (GLMS). RESULTS: The crystal
structures of the enzyme complexed with glutamate and with a competitive
inhibitor, Glu-hydroxamate, have been determined to 1.8 A resolution. The
protein fold has structural homology to other members of the superfamily of
N-terminal nucleophile (Ntn) hydrolases, being a sandwich of antiparallel beta
sheets surrounded by two layers of alpha helices. CONCLUSIONS: The structural
homology between the glutaminase domain of GLMS and that of PRPP
amidotransferase (the only other Ntn amidotransferase whose structure is known)
indicates that they may have diverged from a common ancestor. Cys1 is the
catalytic nucleophile in GLMS, and the nucleophilic character of its thiol group
appears to be increased through general base activation by its own alpha-amino
group. Cys1 can adopt two conformations, one active and one inactive; glutamine
binding locks the residue in a predetermined conformation. We propose that when
a nitrogen acceptor is present Cys1 is kept in the active conformation,
explaining the phenomenon of substrate-induced activation of the enzyme, and
that Arg26 is central in this coupling.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
Figure 5. Active site of the glutaminase domain of GLMS
with the bound product, glutamate, shown in outline. Hydrogen
bonds are indicated by dashed lines. The loop 73-78 is in the
closed conformation. Cys1 and Asn98 are in the inactive
conformation.
|
 |
Figure 6.
Figure 6. Proposed catalytic mechanism of glutamine
hydrolysis by amidotransferases. Residue numbers are those of
GLMS. The nucleophilicity of Cys1 is enhanced by its own free
a-amino group. This interaction is mediated by a bridging water
molecule which serves as a virtual base. Deacylation involves
another water molecule which is activated through the same
mechanism. Residues Asn98 and Gly99 form the oxyanion hole for
the tetrahedral intermediates.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Structure
(1996,
4,
801-810)
copyright 1996.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.Lakomek,
A.Dickmanns,
M.Kettwig,
H.Urlaub,
R.Ficner,
and
T.Lübke
(2009).
Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography.
|
| |
BMC Struct Biol,
9,
56.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.D.Levy,
and
J.B.Pereira-Leal
(2008).
Evolution and dynamics of protein interactions and networks.
|
| |
Curr Opin Struct Biol,
18,
349-357.
|
 |
|
|
|
|
 |
H.Barreteau,
A.Kovac,
A.Boniface,
M.Sova,
S.Gobec,
and
D.Blanot
(2008).
Cytoplasmic steps of peptidoglycan biosynthesis.
|
| |
FEMS Microbiol Rev,
32,
168-207.
|
 |
|
|
|
|
 |
S.Mouilleron,
and
B.Golinelli-Pimpaneau
(2007).
Conformational changes in ammonia-channeling glutamine amidotransferases.
|
| |
Curr Opin Struct Biol,
17,
653-664.
|
 |
|
|
|
|
 |
Y.Wang,
and
H.C.Guo
(2007).
Crystallographic snapshot of a productive glycosylasparaginase-substrate complex.
|
| |
J Mol Biol,
366,
82-92.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Seetharaman,
K.R.Rajashankar,
V.Solorzano,
R.Kniewel,
C.D.Lima,
J.B.Bonanno,
S.K.Burley,
and
S.Swaminathan
(2006).
Crystal structures of two putative phosphoheptose isomerases.
|
| |
Proteins,
63,
1092-1096.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Milewski,
I.Gabriel,
and
J.Olchowy
(2006).
Enzymes of UDP-GlcNAc biosynthesis in yeast.
|
| |
Yeast,
23,
1.
|
 |
|
|
|
|
 |
S.Mouilleron,
M.A.Badet-Denisot,
and
B.Golinelli-Pimpaneau
(2006).
Glutamine binding opens the ammonia channel and activates glucosamine-6P synthase.
|
| |
J Biol Chem,
281,
4404-4412.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.Levitin,
O.Stern,
M.Weiss,
C.Gil-Henn,
R.Ziv,
Z.Prokocimer,
N.I.Smorodinsky,
D.B.Rubinstein,
and
D.H.Wreschner
(2005).
The MUC1 SEA module is a self-cleaving domain.
|
| |
J Biol Chem,
280,
33374-33386.
|
 |
|
|
|
|
 |
J.A.Khan,
B.M.Dunn,
and
L.Tong
(2005).
Crystal structure of human Taspase1, a crucial protease regulating the function of MLL.
|
| |
Structure,
13,
1443-1452.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.H.Lin,
G.W.Chang,
J.Q.Davies,
M.Stacey,
J.Harris,
and
S.Gordon
(2004).
Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif.
|
| |
J Biol Chem,
279,
31823-31832.
|
 |
|
|
|
|
 |
V.M.Coiro,
A.Di Nola,
M.A.Vanoni,
M.Aschi,
A.Coda,
B.Curti,
and
D.Roccatano
(2004).
Molecular dynamics simulation of the interaction between the complex iron-sulfur flavoprotein glutamate synthase and its substrates.
|
| |
Protein Sci,
13,
2979-2991.
|
 |
|
|
|
|
 |
F.Schmitzberger,
M.L.Kilkenny,
C.M.Lobley,
M.E.Webb,
M.Vinkovic,
D.Matak-Vinkovic,
M.Witty,
D.Y.Chirgadze,
A.G.Smith,
C.Abell,
and
T.L.Blundell
(2003).
Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase.
|
| |
EMBO J,
22,
6193-6204.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Wiame,
G.Delpierre,
F.Collard,
and
E.Van Schaftingen
(2002).
Identification of a pathway for the utilization of the Amadori product fructoselysine in Escherichia coli.
|
| |
J Biol Chem,
277,
42523-42529.
|
 |
|
|
|
|
 |
C.Binda,
R.T.Bossi,
S.Wakatsuki,
S.Arzt,
A.Coda,
B.Curti,
M.A.Vanoni,
and
A.Mattevi
(2000).
Cross-talk and ammonia channeling between active centers in the unexpected domain arrangement of glutamate synthase.
|
| |
Structure,
8,
1299-1308.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Bompard-Gilles,
V.Villeret,
G.J.Davies,
L.Fanuel,
B.Joris,
J.M.Frère,
and
J.Van Beeumen
(2000).
A new variant of the Ntn hydrolase fold revealed by the crystal structure of L-aminopeptidase D-ala-esterase/amidase from Ochrobactrum anthropi.
|
| |
Structure,
8,
153-162.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Oinonen,
and
J.Rouvinen
(2000).
Structural comparison of Ntn-hydrolases.
|
| |
Protein Sci,
9,
2329-2337.
|
 |
|
|
|
|
 |
F.Krekel,
A.K.Samland,
P.Macheroux,
N.Amrhein,
and
J.N.Evans
(2000).
Determination of the pKa value of C115 in MurA (UDP-N-acetylglucosamine enolpyruvyltransferase) from Enterobacter cloacae.
|
| |
Biochemistry,
39,
12671-12677.
|
 |
|
|
|
|
 |
T.Nakai,
T.Hasegawa,
E.Yamashita,
M.Yamamoto,
T.Kumasaka,
T.Ueki,
H.Nanba,
Y.Ikenaka,
S.Takahashi,
M.Sato,
and
T.Tsukihara
(2000).
Crystal structure of N-carbamyl-D-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases.
|
| |
Structure,
8,
729-737.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Teplyakov,
G.Obmolova,
M.A.Badet-Denisot,
and
B.Badet
(1999).
The mechanism of sugar phosphate isomerization by glucosamine 6-phosphate synthase.
|
| |
Protein Sci,
8,
596-602.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.G.Schnizer,
S.K.Boehlein,
J.D.Stewart,
N.G.Richards,
and
S.M.Schuster
(1999).
Formation and isolation of a covalent intermediate during the glutaminase reaction of a class II amidotransferase.
|
| |
Biochemistry,
38,
3677-3682.
|
 |
|
|
|
|
 |
N.N.Aronson
(1999).
Aspartylglycosaminuria: biochemistry and molecular biology.
|
| |
Biochim Biophys Acta,
1455,
139-154.
|
 |
|
|
|
|
 |
Q.Xu,
D.Buckley,
C.Guan,
and
H.C.Guo
(1999).
Structural insights into the mechanism of intramolecular proteolysis.
|
| |
Cell,
98,
651-661.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.J.Billington,
A.S.Huggins,
P.A.Johanesen,
P.K.Crellin,
J.K.Cheung,
M.E.Katz,
C.L.Wright,
V.Haring,
and
J.I.Rood
(1999).
Complete nucleotide sequence of the 27-kilobase virulence related locus (vrl) of Dichelobacter nodosus: evidence for extrachromosomal origin.
|
| |
Infect Immun,
67,
1277-1286.
|
 |
|
|
|
|
 |
T.M.Larsen,
S.K.Boehlein,
S.M.Schuster,
N.G.Richards,
J.B.Thoden,
H.M.Holden,
and
I.Rayment
(1999).
Three-dimensional structure of Escherichia coli asparagine synthetase B: a short journey from substrate to product.
|
| |
Biochemistry,
38,
16146-16157.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Teplyakov,
G.Obmolova,
M.A.Badet-Denisot,
B.Badet,
and
I.Polikarpov
(1998).
Involvement of the C terminus in intramolecular nitrogen channeling in glucosamine 6-phosphate synthase: evidence from a 1.6 A crystal structure of the isomerase domain.
|
| |
Structure,
6,
1047-1055.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.R.Muchmore,
J.M.Krahn,
J.H.Kim,
H.Zalkin,
and
J.L.Smith
(1998).
Crystal structure of glutamine phosphoribosylpyrophosphate amidotransferase from Escherichia coli.
|
| |
Protein Sci,
7,
39-51.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.L.Smith
(1998).
Glutamine PRPP amidotransferase: snapshots of an enzyme in action.
|
| |
Curr Opin Struct Biol,
8,
686-694.
|
 |
|
|
|
|
 |
M.Rizzi,
M.Bolognesi,
and
A.Coda
(1998).
A novel deamido-NAD+-binding site revealed by the trapped NAD-adenylate intermediate in the NAD+ synthetase structure.
|
| |
Structure,
6,
1129-1140.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.M.Krahn,
J.H.Kim,
M.R.Burns,
R.J.Parry,
H.Zalkin,
and
J.L.Smith
(1997).
Coupled formation of an amidotransferase interdomain ammonia channel and a phosphoribosyltransferase active site.
|
| |
Biochemistry,
36,
11061-11068.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.K.Boehlein,
E.S.Walworth,
and
S.M.Schuster
(1997).
Identification of cysteine-523 in the aspartate binding site of Escherichia coli asparagine synthetase B.
|
| |
Biochemistry,
36,
10168-10177.
|
 |
|
|
|
|
 |
G.Schmidtke,
R.Kraft,
S.Kostka,
P.Henklein,
C.Frömmel,
J.Löwe,
R.Huber,
P.M.Kloetzel,
and
M.Schmidt
(1996).
Analysis of mammalian 20S proteasome biogenesis: the maturation of beta-subunits is an ordered two-step mechanism involving autocatalysis.
|
| |
EMBO J,
15,
6887-6898.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

