 |
PDBsum entry 2gl9

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D:
E.C.3.5.1.26
- N(4)-(beta-N-acetylglucosaminyl)-L-asparaginase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N4-(beta-N-acetyl-D-glucosaminyl)-L-asparagine + H2O = N-acetyl-beta-D- glucosaminylamine + L-aspartate + H+
|
 |
 |
 |
 |
 |
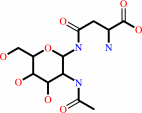 N(4)-(beta-N-acetyl-D-glucosaminyl)-L-asparagine
N(4)-(beta-N-acetyl-D-glucosaminyl)-L-asparagine
|
+
|
H2O
|
=
|
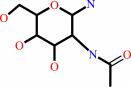 N-acetyl-beta-D- glucosaminylamine
N-acetyl-beta-D- glucosaminylamine
|
+
|
L-aspartate
Bound ligand (Het Group name = )
matches with 80.00% similarity
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 93.33% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
366:82-92
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystallographic snapshot of a productive glycosylasparaginase-substrate complex.
|
|
Y.Wang,
H.C.Guo.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Glycosylasparaginase (GA) plays an important role in asparagine-linked
glycoprotein degradation. A deficiency in the activity of human GA leads to a
lysosomal storage disease named aspartylglycosaminuria. GA belongs to a
superfamily of N-terminal nucleophile hydrolases that autoproteolytically
generate their mature enzymes from inactive single chain protein precursors. The
side-chain of the newly exposed N-terminal residue then acts as a nucleophile
during substrate hydrolysis. By taking advantage of mutant enzyme of
Flavobacterium meningosepticum GA with reduced enzymatic activity, we have
obtained a crystallographic snapshot of a productive complex with its substrate
(NAcGlc-Asn), at 2.0 A resolution. This complex structure provided us an
excellent model for the Michaelis complex to examine the specific contacts
critical for substrate binding and catalysis. Substrate binding induces a
conformational change near the active site of GA. To initiate catalysis, the
side-chain of the N-terminal Thr152 is polarized by the free alpha-amino group
on the same residue, mediated by the side-chain hydroxyl group of Thr170.
Cleavage of the amide bond is then accomplished by a nucleophilic attack at the
carbonyl carbon of the amide linkage in the substrate, leading to the formation
of an acyl-enzyme intermediate through a negatively charged tetrahedral
transition state.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Hydrolysis reaction catalyzed by GA amidase. GA
cleaves the β-N-aspartylglucosylamine bond (indicated by the
bold arrow) of its natural substrate NAcGlc-Asn during
proteolytic processings of asparagine-linked glycoproteins,
resulting in the release of apartic acid and aminoglycan. The
latter product is then further hydrolyzed non-enzymatically to
release ammonia and oligosaccharide. Figure 1. Hydrolysis
reaction catalyzed by GA amidase. GA cleaves the
β-N-aspartylglucosylamine bond (indicated by the bold arrow) of
its natural substrate NAcGlc-Asn during proteolytic processings
of asparagine-linked glycoproteins, resulting in the release of
apartic acid and aminoglycan. The latter product is then further
hydrolyzed non-enzymatically to release ammonia and
oligosaccharide.
|
 |
Figure 4.
Figure 4. Stereo view of the atomic interactions between GA and
substrate. Displayed are interactions between GA and the bound
substrate molecule NAcGlc-Asn at the active site A. The active
side-chain conformation of residue Cys152 is shown in magenta
and the switch between its inactive trans- and active gauche(+)
conformations is indicated by the magenta double arrow.
Nucleophilic attack is indicated by the green straight arrow. A
candidate water molecule to protonate the leaving group is also
shown (W). The green dotted lines indicate possible
hydrogen-bonding interactions between Cys152 and the surrounding
residues. The blue dotted lines denote other hydrogen bonds
involved in enzyme–substrate binding. Also shown is a hydrogen
bond (a black dotted line) between side-chains of Trp11 and
Thr203. Key active site residues are shown by atom type: yellow
for carbon, blue for nitrogen, red for oxygen, and green for the
Cys152 sulfur atom. The salt bridge is indicated by the
positive and the negative charges. Figure 4. Stereo view of
the atomic interactions between GA and substrate. Displayed are
interactions between GA and the bound substrate molecule
NAcGlc-Asn at the active site A. The active side-chain
conformation of residue Cys152 is shown in magenta and the
switch between its inactive trans- and active gauche(+)
conformations is indicated by the magenta double arrow.
Nucleophilic attack is indicated by the green straight arrow. A
candidate water molecule to protonate the leaving group is also
shown (W). The green dotted lines indicate possible
hydrogen-bonding interactions between Cys152 and the surrounding
residues. The blue dotted lines denote other hydrogen bonds
involved in enzyme–substrate binding. Also shown is a hydrogen
bond (a black dotted line) between side-chains of Trp11 and
Thr203. Key active site residues are shown by atom type: yellow
for carbon, blue for nitrogen, red for oxygen, and green for the
Cys152 sulfur atom. The salt bridge is indicated by the positive
and the negative charges.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Elsevier:
J Mol Biol
(2007,
366,
82-92)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.Sun,
and
H.C.Guo
(2008).
Structural constraints on autoprocessing of the human nucleoporin Nup98.
|
| |
Protein Sci,
17,
494-505.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |
|

