 |
PDBsum entry 1ecc

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.14
- amidophosphoribosyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5-phospho-beta-D-ribosylamine + L-glutamate + diphosphate = 5-phospho- alpha-D-ribose 1-diphosphate + L-glutamine + H2O
|
 |
 |
 |
 |
 |
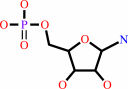 5-phospho-beta-D-ribosylamine
5-phospho-beta-D-ribosylamine
|
+
|
 L-glutamate
L-glutamate
|
+
|
diphosphate
Bound ligand (Het Group name = )
matches with 81.82% similarity
|
=
|
5-phospho- alpha-D-ribose 1-diphosphate
Bound ligand (Het Group name = )
matches with 91.30% similarity
|
+
|
 L-glutamine
L-glutamine
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
36:11061-11068
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
Coupled formation of an amidotransferase interdomain ammonia channel and a phosphoribosyltransferase active site.
|
|
J.M.Krahn,
J.H.Kim,
M.R.Burns,
R.J.Parry,
H.Zalkin,
J.L.Smith.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Activation of gluatmine phosphoribosylpyrophosphate (RPPP) amidotransferase
(GPATase) by binding of a PRPP substrate analog results in the formation of a 20
A channel connecting the active site for glutamine hydrolysis in one domain with
the PRPP site in a second domain. This solvent-inaccessible channel permits
transfer of the NH3 intermediate between the two active sites. Tunneling of NH3
may be a common mechanism for glutamine amidotransferase-catalyzed nitrogen
transfer and for coordination of catalysis at two distinct active sites in
complex enzymes. The 2.4 A crystal structure of the active conformer of GPATase
also provides the first description of an intact active site for the
phosphoribosyltransferase (PRTase) family of nucleotide synthesis and salvage
enzymes. Chemical assistance to catalysis is provided primarily by the substrate
and secondarily by the enzyme in the proposed structure-based mechanism.
Different catalytic and inhibitory modes of divalent cation binding to the
PRTase active site are revealed in the active conformer of the enzyme and in a
feedback-inhibited GMP complex.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.M.Lipchock,
and
J.P.Loria
(2010).
Nanometer propagation of millisecond motions in V-type allostery.
|
| |
Structure,
18,
1596-1607.
|
 |
|
|
|
|
 |
K.Hölsch,
and
D.Weuster-Botz
(2010).
Enantioselective reduction of prochiral ketones by engineered bifunctional fusion proteins.
|
| |
Biotechnol Appl Biochem,
56,
131-140.
|
 |
|
|
|
|
 |
L.Lund,
Y.Fan,
Q.Shao,
Y.Q.Gao,
and
F.M.Raushel
(2010).
Carbamate transport in carbamoyl phosphate synthetase: a theoretical and experimental investigation.
|
| |
J Am Chem Soc,
132,
3870-3878.
|
 |
|
|
|
|
 |
R.Takahashi,
S.Nakamura,
T.Nakazawa,
K.Minoura,
T.Yoshida,
Y.Nishi,
Y.Kobayashi,
and
T.Ohkubo
(2010).
Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase.
|
| |
J Biochem,
147,
95.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Lipchock,
and
J.P.Loria
(2009).
Millisecond dynamics in the allosteric enzyme imidazole glycerol phosphate synthase (IGPS) from Thermotoga maritima.
|
| |
J Biomol NMR,
45,
73-84.
|
 |
|
|
|
|
 |
R.L.Switzer
(2009).
Discoveries in bacterial nucleotide metabolism.
|
| |
J Biol Chem,
284,
6585-6594.
|
 |
|
|
|
|
 |
Y.Fan,
L.Lund,
Q.Shao,
Y.Q.Gao,
and
F.M.Raushel
(2009).
A combined theoretical and experimental study of the ammonia tunnel in carbamoyl phosphate synthetase.
|
| |
J Am Chem Soc,
131,
10211-10219.
|
 |
|
|
|
|
 |
W.M.Patrick,
and
I.Matsumura
(2008).
A study in molecular contingency: glutamine phosphoribosylpyrophosphate amidotransferase is a promiscuous and evolvable phosphoribosylanthranilate isomerase.
|
| |
J Mol Biol,
377,
323-336.
|
 |
|
|
|
|
 |
Y.Zhang,
M.Morar,
and
S.E.Ealick
(2008).
Structural biology of the purine biosynthetic pathway.
|
| |
Cell Mol Life Sci,
65,
3699-3724.
|
 |
|
|
|
|
 |
S.Mouilleron,
and
B.Golinelli-Pimpaneau
(2007).
Conformational changes in ammonia-channeling glutamine amidotransferases.
|
| |
Curr Opin Struct Biol,
17,
653-664.
|
 |
|
|
|
|
 |
N.G.Richards,
and
M.S.Kilberg
(2006).
Asparagine synthetase chemotherapy.
|
| |
Annu Rev Biochem,
75,
629-654.
|
 |
|
|
|
|
 |
S.Mouilleron,
M.A.Badet-Denisot,
and
B.Golinelli-Pimpaneau
(2006).
Glutamine binding opens the ammonia channel and activates glucosamine-6P synthase.
|
| |
J Biol Chem,
281,
4404-4412.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Zhu,
J.W.Burgner,
E.Harms,
B.R.Belitsky,
and
J.L.Smith
(2005).
A new arrangement of (beta/alpha)8 barrels in the synthase subunit of PLP synthase.
|
| |
J Biol Chem,
280,
27914-27923.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Chander,
K.M.Halbig,
J.K.Miller,
C.J.Fields,
H.K.Bonner,
G.K.Grabner,
R.L.Switzer,
and
J.L.Smith
(2005).
Structure of the nucleotide complex of PyrR, the pyr attenuation protein from Bacillus caldolyticus, suggests dual regulation by pyrimidine and purine nucleotides.
|
| |
J Bacteriol,
187,
1773-1782.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Mitani,
X.Meng,
Y.Kamagata,
and
T.Tamura
(2005).
Characterization of LtsA from Rhodococcus erythropolis, an enzyme with glutamine amidotransferase activity.
|
| |
J Bacteriol,
187,
2582-2591.
|
 |
|
|
|
|
 |
F.A.Lunn,
and
S.L.Bearne
(2004).
Alternative substrates for wild-type and L109A E. coli CTP synthases: kinetic evidence for a constricted ammonia tunnel.
|
| |
Eur J Biochem,
271,
4204-4212.
|
 |
|
|
|
|
 |
J.D.Lawson,
E.Pate,
I.Rayment,
and
R.G.Yount
(2004).
Molecular dynamics analysis of structural factors influencing back door pi release in myosin.
|
| |
Biophys J,
86,
3794-3803.
|
 |
|
|
|
|
 |
L.Salomonsson,
A.Lee,
R.B.Gennis,
and
P.Brzezinski
(2004).
A single-amino-acid lid renders a gas-tight compartment within a membrane-bound transporter.
|
| |
Proc Natl Acad Sci U S A,
101,
11617-11621.
|
 |
|
|
|
|
 |
A.K.Bera,
J.Zhu,
H.Zalkin,
and
J.L.Smith
(2003).
Functional dissection of the Bacillus subtilis pur operator site.
|
| |
J Bacteriol,
185,
4099-4109.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Saeed-Kothe,
and
S.G.Powers-Lee
(2003).
Gain of glutaminase function in mutants of the ammonia-specific frog carbamoyl phosphate synthetase.
|
| |
J Biol Chem,
278,
26722-26726.
|
 |
|
|
|
|
 |
B.A.Manjasetty,
J.Powlowski,
and
A.Vrielink
(2003).
Crystal structure of a bifunctional aldolase-dehydrogenase: sequestering a reactive and volatile intermediate.
|
| |
Proc Natl Acad Sci U S A,
100,
6992-6997.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.K.Grabner,
and
R.L.Switzer
(2003).
Kinetic studies of the uracil phosphoribosyltransferase reaction catalyzed by the Bacillus subtilis pyrimidine attenuation regulatory protein PyrR.
|
| |
J Biol Chem,
278,
6921-6927.
|
 |
|
|
|
|
 |
R.Amaro,
E.Tajkhorshid,
and
Z.Luthey-Schulten
(2003).
Developing an energy landscape for the novel function of a (beta/alpha)8 barrel: ammonia conduction through HisF.
|
| |
Proc Natl Acad Sci U S A,
100,
7599-7604.
|
 |
|
|
|
|
 |
S.C.Sinha,
J.Krahn,
B.S.Shin,
D.R.Tomchick,
H.Zalkin,
and
J.L.Smith
(2003).
The purine repressor of Bacillus subtilis: a novel combination of domains adapted for transcription regulation.
|
| |
J Bacteriol,
185,
4087-4098.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Douangamath,
M.Walker,
S.Beismann-Driemeyer,
M.C.Vega-Fernandez,
R.Sterner,
and
M.Wilmanns
(2002).
Structural evidence for ammonia tunneling across the (beta alpha)(8) barrel of the imidazole glycerol phosphate synthase bienzyme complex.
|
| |
Structure,
10,
185-193.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Kadziola,
J.Neuhard,
and
S.Larsen
(2002).
Structure of product-bound Bacillus caldolyticus uracil phosphoribosyltransferase confirms ordered sequential substrate binding.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
936-945.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.B.Thoden,
X.Huang,
F.M.Raushel,
and
H.M.Holden
(2002).
Carbamoyl-phosphate synthetase. Creation of an escape route for ammonia.
|
| |
J Biol Chem,
277,
39722-39727.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.G.Bulock,
G.P.Beardsley,
and
K.S.Anderson
(2002).
The kinetic mechanism of the human bifunctional enzyme ATIC (5-amino-4-imidazolecarboxamide ribonucleotide transformylase/inosine 5'-monophosphate cyclohydrolase). A surprising lack of substrate channeling.
|
| |
J Biol Chem,
277,
22168-22174.
|
 |
|
|
|
|
 |
M.A.Schumacher,
C.J.Bashor,
M.H.Song,
K.Otsu,
S.Zhu,
R.J.Parry,
B.Ullman,
and
R.G.Brennan
(2002).
The structural mechanism of GTP stabilized oligomerization and catalytic activation of the Toxoplasma gondii uracil phosphoribosyltransferase.
|
| |
Proc Natl Acad Sci U S A,
99,
78-83.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.H.van den Heuvel,
D.Ferrari,
R.T.Bossi,
S.Ravasio,
B.Curti,
M.A.Vanoni,
F.J.Florencio,
and
A.Mattevi
(2002).
Structural studies on the synchronization of catalytic centers in glutamate synthase.
|
| |
J Biol Chem,
277,
24579-24583.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.A.Eriksen,
A.Kadziola,
and
S.Larsen
(2002).
Binding of cations in Bacillus subtilis phosphoribosyldiphosphate synthetase and their role in catalysis.
|
| |
Protein Sci,
11,
271-279.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Bergner,
J.Günther,
M.Hendlich,
G.Klebe,
and
M.Verdonk
(2001).
Use of Relibase for retrieving complex three-dimensional interaction patterns including crystallographic packing effects.
|
| |
Biopolymers,
61,
99.
|
 |
|
|
|
|
 |
I.Nobeli,
R.A.Laskowski,
W.S.Valdar,
and
J.M.Thornton
(2001).
On the molecular discrimination between adenine and guanine by proteins.
|
| |
Nucleic Acids Res,
29,
4294-4309.
|
 |
|
|
|
|
 |
T.J.Klem,
Y.Chen,
and
V.J.Davisson
(2001).
Subunit interactions and glutamine utilization by Escherichia coli imidazole glycerol phosphate synthase.
|
| |
J Bacteriol,
183,
989-996.
|
 |
|
|
|
|
 |
X.Huang,
H.M.Holden,
and
F.M.Raushel
(2001).
Channeling of substrates and intermediates in enzyme-catalyzed reactions.
|
| |
Annu Rev Biochem,
70,
149-180.
|
 |
|
|
|
|
 |
A.K.Bera,
J.L.Smith,
and
H.Zalkin
(2000).
Dual role for the glutamine phosphoribosylpyrophosphate amidotransferase ammonia channel. Interdomain signaling and intermediate channeling.
|
| |
J Biol Chem,
275,
7975-7979.
|
 |
|
|
|
|
 |
A.K.Bera,
S.Chen,
J.L.Smith,
and
H.Zalkin
(2000).
Temperature-dependent function of the glutamine phosphoribosylpyrophosphate amidotransferase ammonia channel and coupling with glycinamide ribonucleotide synthetase in a hyperthermophile.
|
| |
J Bacteriol,
182,
3734-3739.
|
 |
|
|
|
|
 |
C.Binda,
R.T.Bossi,
S.Wakatsuki,
S.Arzt,
A.Coda,
B.Curti,
M.A.Vanoni,
and
A.Mattevi
(2000).
Cross-talk and ammonia channeling between active centers in the unexpected domain arrangement of glutamate synthase.
|
| |
Structure,
8,
1299-1308.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Kohls,
T.Sulea,
E.O.Purisima,
R.E.MacKenzie,
and
A.Vrielink
(2000).
The crystal structure of the formiminotransferase domain of formiminotransferase-cyclodeaminase: implications for substrate channeling in a bifunctional enzyme.
|
| |
Structure,
8,
35-46.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Reumann
(2000).
The structural properties of plant peroxisomes and their metabolic significance.
|
| |
Biol Chem,
381,
639-648.
|
 |
|
|
|
|
 |
T.J.Kappock,
S.E.Ealick,
and
J.Stubbe
(2000).
Modular evolution of the purine biosynthetic pathway.
|
| |
Curr Opin Chem Biol,
4,
567-572.
|
 |
|
|
|
|
 |
W.Shi,
N.R.Munagala,
C.C.Wang,
C.M.Li,
P.C.Tyler,
R.H.Furneaux,
C.Grubmeyer,
V.L.Schramm,
and
S.C.Almo
(2000).
Crystal structures of Giardia lamblia guanine phosphoribosyltransferase at 1.75 A(,).
|
| |
Biochemistry,
39,
6781-6790.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
X.Huang,
and
F.M.Raushel
(2000).
An engineered blockage within the ammonia tunnel of carbamoyl phosphate synthetase prevents the use of glutamine as a substrate but not ammonia.
|
| |
Biochemistry,
39,
3240-3247.
|
 |
|
|
|
|
 |
A.K.Bera,
S.Chen,
J.L.Smith,
and
H.Zalkin
(1999).
Interdomain signaling in glutamine phosphoribosylpyrophosphate amidotransferase.
|
| |
J Biol Chem,
274,
36498-36504.
|
 |
|
|
|
|
 |
C.L.Phillips,
B.Ullman,
R.G.Brennan,
and
C.P.Hill
(1999).
Crystal structures of adenine phosphoribosyltransferase from Leishmania donovani.
|
| |
EMBO J,
18,
3533-3545.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Li,
T.J.Kappock,
J.Stubbe,
T.M.Weaver,
and
S.E.Ealick
(1999).
X-ray crystal structure of aminoimidazole ribonucleotide synthetase (PurM), from the Escherichia coli purine biosynthetic pathway at 2.5 A resolution.
|
| |
Structure,
7,
1155-1166.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Lundegaard,
and
K.F.Jensen
(1999).
Kinetic mechanism of uracil phosphoribosyltransferase from Escherichia coli and catalytic importance of the conserved proline in the PRPP binding site.
|
| |
Biochemistry,
38,
3327-3334.
|
 |
|
|
|
|
 |
E.W.Miles,
S.Rhee,
and
D.R.Davies
(1999).
The molecular basis of substrate channeling.
|
| |
J Biol Chem,
274,
12193-12196.
|
 |
|
|
|
|
 |
F.M.Raushel,
J.B.Thoden,
and
H.M.Holden
(1999).
The amidotransferase family of enzymes: molecular machines for the production and delivery of ammonia.
|
| |
Biochemistry,
38,
7891-7899.
|
 |
|
|
|
|
 |
G.K.Balendiran,
J.A.Molina,
Y.Xu,
J.Torres-Martinez,
R.Stevens,
P.J.Focia,
A.E.Eakin,
J.C.Sacchettini,
and
S.P.Craig
(1999).
Ternary complex structure of human HGPRTase, PRPP, Mg2+, and the inhibitor HPP reveals the involvement of the flexible loop in substrate binding.
|
| |
Protein Sci,
8,
1023-1031.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.P.Wang,
C.Lundegaard,
K.F.Jensen,
and
C.Grubmeyer
(1999).
Kinetic mechanism of OMP synthase: a slow physical step following group transfer limits catalytic rate.
|
| |
Biochemistry,
38,
275-283.
|
 |
|
|
|
|
 |
H.G.Schnizer,
S.K.Boehlein,
J.D.Stewart,
N.G.Richards,
and
S.M.Schuster
(1999).
Formation and isolation of a covalent intermediate during the glutaminase reaction of a class II amidotransferase.
|
| |
Biochemistry,
38,
3677-3682.
|
 |
|
|
|
|
 |
I.I.Mathews,
T.J.Kappock,
J.Stubbe,
and
S.E.Ealick
(1999).
Crystal structure of Escherichia coli PurE, an unusual mutase in the purine biosynthetic pathway.
|
| |
Structure,
7,
1395-1406.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.P.Page,
N.R.Munagala,
and
C.C.Wang
(1999).
Point mutations in the guanine phosphoribosyltransferase from Giardia lamblia modulate pyrophosphate binding and enzyme catalysis.
|
| |
Eur J Biochem,
259,
565-571.
|
 |
|
|
|
|
 |
M.K.Geck,
and
J.F.Kirsch
(1999).
A novel, definitive test for substrate channeling illustrated with the aspartate aminotransferase/malate dehydrogenase system.
|
| |
Biochemistry,
38,
8032-8037.
|
 |
|
|
|
|
 |
P.Rappu,
B.S.Shin,
H.Zalkin,
and
P.Mäntsälä
(1999).
A role for a highly conserved protein of unknown function in regulation of Bacillus subtilis purA by the purine repressor.
|
| |
J Bacteriol,
181,
3810-3815.
|
 |
|
|
|
|
 |
S.Li,
J.L.Smith,
and
H.Zalkin
(1999).
Mutational analysis of Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase propeptide processing.
|
| |
J Bacteriol,
181,
1403-1408.
|
 |
|
|
|
|
 |
T.M.Larsen,
S.K.Boehlein,
S.M.Schuster,
N.G.Richards,
J.B.Thoden,
H.M.Holden,
and
I.Rayment
(1999).
Three-dimensional structure of Escherichia coli asparagine synthetase B: a short journey from substrate to product.
|
| |
Biochemistry,
38,
16146-16157.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Teplyakov,
G.Obmolova,
M.A.Badet-Denisot,
B.Badet,
and
I.Polikarpov
(1998).
Involvement of the C terminus in intramolecular nitrogen channeling in glucosamine 6-phosphate synthase: evidence from a 1.6 A crystal structure of the isomerase domain.
|
| |
Structure,
6,
1047-1055.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.O.Bachmann,
R.Li,
and
C.A.Townsend
(1998).
beta-Lactam synthetase: a new biosynthetic enzyme.
|
| |
Proc Natl Acad Sci U S A,
95,
9082-9086.
|
 |
|
|
|
|
 |
D.R.Tomchick,
R.J.Turner,
R.L.Switzer,
and
J.L.Smith
(1998).
Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase.
|
| |
Structure,
6,
337-350.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.M.Raushel,
J.B.Thoden,
G.D.Reinhart,
and
H.M.Holden
(1998).
Carbamoyl phosphate synthetase: a crooked path from substrates to products.
|
| |
Curr Opin Chem Biol,
2,
624-632.
|
 |
|
|
|
|
 |
H.M.Holden,
J.B.Thoden,
and
F.M.Raushel
(1998).
Carbamoyl phosphate synthetase: a tunnel runs through it.
|
| |
Curr Opin Struct Biol,
8,
679-685.
|
 |
|
|
|
|
 |
J.L.Smith
(1998).
Glutamine PRPP amidotransferase: snapshots of an enzyme in action.
|
| |
Curr Opin Struct Biol,
8,
686-694.
|
 |
|
|
|
|
 |
M.A.Schumacher,
D.Carter,
D.M.Scott,
D.S.Roos,
B.Ullman,
and
R.G.Brennan
(1998).
Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
|
| |
EMBO J,
17,
3219-3232.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Rizzi,
M.Bolognesi,
and
A.Coda
(1998).
A novel deamido-NAD+-binding site revealed by the trapped NAD-adenylate intermediate in the NAD+ synthetase structure.
|
| |
Structure,
6,
1129-1140.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.R.Munagala,
M.S.Chin,
and
C.C.Wang
(1998).
Steady-state kinetics of the hypoxanthine-guanine-xanthine phosphoribosyltransferase from Tritrichomonas foetus: the role of threonine-47.
|
| |
Biochemistry,
37,
4045-4051.
|
 |
|
|
|
|
 |
P.J.Focia,
S.P.Craig,
and
A.E.Eakin
(1998).
Approaching the transition state in the crystal structure of a phosphoribosyltransferase.
|
| |
Biochemistry,
37,
17120-17127.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.J.Focia,
S.P.Craig,
R.Nieves-Alicea,
R.J.Fletterick,
and
A.E.Eakin
(1998).
A 1.4 A crystal structure for the hypoxanthine phosphoribosyltransferase of Trypanosoma cruzi.
|
| |
Biochemistry,
37,
15066-15075.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
V.Sharma,
C.Grubmeyer,
and
J.C.Sacchettini
(1998).
Crystal structure of quinolinic acid phosphoribosyltransferase from Mmycobacterium tuberculosis: a potential TB drug target.
|
| |
Structure,
6,
1587-1599.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.Wang,
T.J.Kappock,
J.Stubbe,
and
S.E.Ealick
(1998).
X-ray crystal structure of glycinamide ribonucleotide synthetase from Escherichia coli.
|
| |
Biochemistry,
37,
15647-15662.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Xu,
and
C.Grubmeyer
(1998).
Catalysis in human hypoxanthine-guanine phosphoribosyltransferase: Asp 137 acts as a general acid/base.
|
| |
Biochemistry,
37,
4114-4124.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

