 |
PDBsum entry 1gpm

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase (glutamine amidotransferase)
|
PDB id
|
|

|

|
1gpm
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.5.2
- Gmp synthase (glutamine-hydrolyzing).
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP and GMP Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
XMP + L-glutamine + ATP + H2O = GMP + L-glutamate + AMP + diphosphate + 2 H+
|
 |
 |
 |
 |
 |
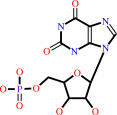 XMP
XMP
|
+
|
 L-glutamine
L-glutamine
|
+
|
 ATP
ATP
|
+
|
H2O
|
=
|
GMP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
L-glutamate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
+
|
2
×
H(+)
Bound ligand (Het Group name = )
matches with 64.29% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Nat Struct Biol
3:74-86
(1996)
|
|
PubMed id:
|
|
|
|

|
| |
|
The crystal structure of GMP synthetase reveals a novel catalytic triad and is a structural paradigm for two enzyme families.
|
|
J.J.Tesmer,
T.J.Klem,
M.L.Deras,
V.J.Davisson,
J.L.Smith.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of GMP synthetase serves as a prototype for two families
of metabolic enzymes. The Class I glutamine amidotransferase domain of GMP
synthetase is found in related enzymes of the purine, pyrimidine, tryptophan,
arginine, histidine and folic acid biosynthetic pathways. This domain includes a
conserved Cys-His-Glu triad and is representative of a new family of enzymes
that use a catalytic triad for enzymatic hydrolysis. The structure and conserved
sequence fingerprint of the nucleotide-binding site in a second domain of GMP
synthetase are common to a family of ATP pyrophosphatases, including NAD
synthetase, asparagine synthetase and argininosuccinate synthetase.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
F.Sarkari,
T.Sanchez-Alcaraz,
S.Wang,
M.N.Holowaty,
Y.Sheng,
and
L.Frappier
(2009).
EBNA1-mediated recruitment of a histone H2B deubiquitylating complex to the Epstein-Barr virus latent origin of DNA replication.
|
| |
PLoS Pathog,
5,
e1000624.
|
 |
|
|
|
|
 |
N.LaRonde-LeBlanc,
M.Resto,
and
B.Gerratana
(2009).
Regulation of active site coupling in glutamine-dependent NAD(+) synthetase.
|
| |
Nat Struct Mol Biol,
16,
421-429.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Iwata-Reuyl
(2008).
An embarrassment of riches: the enzymology of RNA modification.
|
| |
Curr Opin Chem Biol,
12,
126-133.
|
 |
|
|
|
|
 |
E.J.Hart,
and
S.G.Powers-Lee
(2008).
Mutation analysis of carbamoyl phosphate synthetase: does the structurally conserved glutamine amidotransferase triad act as a functional dyad?
|
| |
Protein Sci,
17,
1120-1128.
|
 |
|
|
|
|
 |
Y.Ikeuchi,
K.Kitahara,
and
T.Suzuki
(2008).
The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon.
|
| |
EMBO J,
27,
2194-2203.
|
 |
|
|
|
|
 |
E.L.Ang,
J.P.Obbard,
and
H.Zhao
(2007).
Probing the molecular determinants of aniline dioxygenase substrate specificity by saturation mutagenesis.
|
| |
FEBS J,
274,
928-939.
|
 |
|
|
|
|
 |
J.R.Davies,
R.M.Jackson,
K.V.Mardia,
and
C.C.Taylor
(2007).
The Poisson Index: a new probabilistic model for protein ligand binding site similarity.
|
| |
Bioinformatics,
23,
3001-3008.
|
 |
|
|
|
|
 |
M.J.Wagemaker,
D.C.Eastwood,
C.van der Drift,
M.S.Jetten,
K.Burton,
L.J.Van Griensven,
and
H.J.Op den Camp
(2007).
Argininosuccinate synthetase and argininosuccinate lyase: two ornithine cycle enzymes from Agaricus bisporus.
|
| |
Mycol Res,
111,
493-502.
|
 |
|
|
|
|
 |
M.Kuratani,
Y.Yoshikawa,
Y.Bessho,
K.Higashijima,
T.Ishii,
R.Shibata,
S.Takahashi,
K.Yutani,
and
S.Yokoyama
(2007).
Structural basis of the initial binding of tRNA(Ile) lysidine synthetase TilS with ATP and L-lysine.
|
| |
Structure,
15,
1642-1653.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Rodriguez-Suarez,
D.Xu,
K.Veillette,
J.Davison,
S.Sillaots,
S.Kauffman,
W.Hu,
J.Bowman,
N.Martel,
S.Trosok,
H.Wang,
L.Zhang,
L.Y.Huang,
Y.Li,
F.Rahkhoodaee,
T.Ransom,
D.Gauvin,
C.Douglas,
P.Youngman,
J.Becker,
B.Jiang,
and
T.Roemer
(2007).
Mechanism-of-action determination of GMP synthase inhibitors and target validation in Candida albicans and Aspergillus fumigatus.
|
| |
Chem Biol,
14,
1163-1175.
|
 |
|
|
|
|
 |
S.Mouilleron,
and
B.Golinelli-Pimpaneau
(2007).
Conformational changes in ammonia-channeling glutamine amidotransferases.
|
| |
Curr Opin Struct Biol,
17,
653-664.
|
 |
|
|
|
|
 |
J.Chartron,
K.S.Carroll,
C.Shiau,
H.Gao,
J.A.Leary,
C.R.Bertozzi,
and
C.D.Stout
(2006).
Substrate recognition, protein dynamics, and iron-sulfur cluster in Pseudomonas aeruginosa adenosine 5'-phosphosulfate reductase.
|
| |
J Mol Biol,
364,
152-169.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.D.Mougous,
D.H.Lee,
S.C.Hubbard,
M.W.Schelle,
D.J.Vocadlo,
J.M.Berger,
and
C.R.Bertozzi
(2006).
Molecular basis for G protein control of the prokaryotic ATP sulfurylase.
|
| |
Mol Cell,
21,
109-122.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.L.Abbott,
J.M.Newell,
C.M.Lightcap,
M.E.Olanich,
D.T.Loughlin,
M.A.Weller,
G.Lam,
S.Pollack,
and
W.A.Patton
(2006).
The effects of removing the GAT domain from E. coli GMP synthetase.
|
| |
Protein J,
25,
483-491.
|
 |
|
|
|
|
 |
M.Gengenbacher,
T.B.Fitzpatrick,
T.Raschle,
K.Flicker,
I.Sinning,
S.Müller,
P.Macheroux,
I.Tews,
and
B.Kappes
(2006).
Vitamin B6 biosynthesis by the malaria parasite Plasmodium falciparum: biochemical and structural insights.
|
| |
J Biol Chem,
281,
3633-3641.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.G.Richards,
and
M.S.Kilberg
(2006).
Asparagine synthetase chemotherapy.
|
| |
Annu Rev Biochem,
75,
629-654.
|
 |
|
|
|
|
 |
N.Shigi,
Y.Sakaguchi,
T.Suzuki,
and
K.Watanabe
(2006).
Identification of two tRNA thiolation genes required for cell growth at extremely high temperatures.
|
| |
J Biol Chem,
281,
14296-14306.
|
 |
|
|
|
|
 |
T.Numata,
Y.Ikeuchi,
S.Fukai,
T.Suzuki,
and
O.Nureki
(2006).
Snapshots of tRNA sulphuration via an adenylated intermediate.
|
| |
Nature,
442,
419-424.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Curis,
I.Nicolis,
C.Moinard,
S.Osowska,
N.Zerrouk,
S.Bénazeth,
and
L.Cynober
(2005).
Almost all about citrulline in mammals.
|
| |
Amino Acids,
29,
177-205.
|
 |
|
|
|
|
 |
J.Zhu,
J.W.Burgner,
E.Harms,
B.R.Belitsky,
and
J.L.Smith
(2005).
A new arrangement of (beta/alpha)8 barrels in the synthase subunit of PLP synthase.
|
| |
J Biol Chem,
280,
27914-27923.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Nakanishi,
S.Fukai,
Y.Ikeuchi,
A.Soma,
Y.Sekine,
T.Suzuki,
and
O.Nureki
(2005).
Structural basis for lysidine formation by ATP pyrophosphatase accompanied by a lysine-specific loop and a tRNA-recognition domain.
|
| |
Proc Natl Acad Sci U S A,
102,
7487-7492.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.S.Almeida,
T.Herrmann,
W.Peti,
I.A.Wilson,
and
K.Wüthrich
(2005).
NMR structure of the conserved hypothetical protein TM0487 from Thermotoga maritima: implications for 216 homologous DUF59 proteins.
|
| |
Protein Sci,
14,
2880-2886.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Umeda,
T.Suzuki,
M.Yukawa,
Y.Ohya,
H.Shindo,
K.Watanabe,
and
T.Suzuki
(2005).
Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases.
|
| |
J Biol Chem,
280,
1613-1624.
|
 |
|
|
|
|
 |
R.E.Amaro,
R.S.Myers,
V.J.Davisson,
and
Z.A.Luthey-Schulten
(2005).
Structural elements in IGP synthase exclude water to optimize ammonia transfer.
|
| |
Biophys J,
89,
475-487.
|
 |
|
|
|
|
 |
Y.Ikeuchi,
A.Soma,
T.Ote,
J.Kato,
Y.Sekine,
and
T.Suzuki
(2005).
molecular mechanism of lysidine synthesis that determines tRNA identity and codon recognition.
|
| |
Mol Cell,
19,
235-246.
|
 |
|
|
|
|
 |
Y.Mitani,
X.Meng,
Y.Kamagata,
and
T.Tamura
(2005).
Characterization of LtsA from Rhodococcus erythropolis, an enzyme with glutamine amidotransferase activity.
|
| |
J Bacteriol,
187,
2582-2591.
|
 |
|
|
|
|
 |
F.A.Lunn,
and
S.L.Bearne
(2004).
Alternative substrates for wild-type and L109A E. coli CTP synthases: kinetic evidence for a constricted ammonia tunnel.
|
| |
Eur J Biochem,
271,
4204-4212.
|
 |
|
|
|
|
 |
J.A.Bauer,
E.M.Bennett,
T.P.Begley,
and
S.E.Ealick
(2004).
Three-dimensional structure of YaaE from Bacillus subtilis, a glutaminase implicated in pyridoxal-5'-phosphate biosynthesis.
|
| |
J Biol Chem,
279,
2704-2711.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.A.Endrizzi,
H.Kim,
P.M.Anderson,
and
E.P.Baldwin
(2004).
Crystal structure of Escherichia coli cytidine triphosphate synthetase, a nucleotide-regulated glutamine amidotransferase/ATP-dependent amidoligase fusion protein and homologue of anticancer and antiparasitic drug targets.
|
| |
Biochemistry,
43,
6447-6463.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.D.Lawson,
E.Pate,
I.Rayment,
and
R.G.Yount
(2004).
Molecular dynamics analysis of structural factors influencing back door pi release in myosin.
|
| |
Biophys J,
86,
3794-3803.
|
 |
|
|
|
|
 |
M.A.Wilson,
C.V.St Amour,
J.L.Collins,
D.Ringe,
and
G.A.Petsko
(2004).
The 1.8-A resolution crystal structure of YDR533Cp from Saccharomyces cerevisiae: a member of the DJ-1/ThiJ/PfpI superfamily.
|
| |
Proc Natl Acad Sci U S A,
101,
1531-1536.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Goto,
R.Omi,
N.Nakagawa,
I.Miyahara,
and
K.Hirotsu
(2004).
Crystal structures of CTP synthetase reveal ATP, UTP, and glutamine binding sites.
|
| |
Structure,
12,
1413-1423.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Soma,
Y.Ikeuchi,
S.Kanemasa,
K.Kobayashi,
N.Ogasawara,
T.Ote,
J.Kato,
K.Watanabe,
Y.Sekine,
and
T.Suzuki
(2003).
An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA.
|
| |
Mol Cell,
12,
689-698.
|
 |
|
|
|
|
 |
D.E.Pilloff,
and
T.S.Leyh
(2003).
Allosteric and catalytic functions of the PPi-binding motif in the ATP sulfurylase-GTPase system.
|
| |
J Biol Chem,
278,
50435-50441.
|
 |
|
|
|
|
 |
D.Simard,
K.A.Hewitt,
F.Lunn,
A.Iyengar,
and
S.L.Bearne
(2003).
Limited proteolysis of Escherichia coli cytidine 5'-triphosphate synthase. Identification of residues required for CTP formation and GTP-dependent activation of glutamine hydrolysis.
|
| |
Eur J Biochem,
270,
2195-2206.
|
 |
|
|
|
|
 |
K.Cox,
T.Watson,
P.Soultanas,
and
J.D.Hirst
(2003).
Molecular dynamics simulations of a helicase.
|
| |
Proteins,
52,
254-262.
|
 |
|
|
|
|
 |
K.Honbou,
N.N.Suzuki,
M.Horiuchi,
T.Niki,
T.Taira,
H.Ariga,
and
F.Inagaki
(2003).
The crystal structure of DJ-1, a protein related to male fertility and Parkinson's disease.
|
| |
J Biol Chem,
278,
31380-31384.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Goto,
R.Omi,
I.Miyahara,
M.Sugahara,
and
K.Hirotsu
(2003).
Structures of argininosuccinate synthetase in enzyme-ATP substrates and enzyme-AMP product forms: stereochemistry of the catalytic reaction.
|
| |
J Biol Chem,
278,
22964-22971.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Hara,
K.Yamada,
M.Terashima,
H.Osago,
M.Shimoyama,
and
M.Tsuchiya
(2003).
Molecular identification of human glutamine- and ammonia-dependent NAD synthetases. Carbon-nitrogen hydrolase domain confers glutamine dependency.
|
| |
J Biol Chem,
278,
10914-10921.
|
 |
|
|
|
|
 |
P.M.Quigley,
K.Korotkov,
F.Baneyx,
and
W.G.Hol
(2003).
The 1.6-A crystal structure of the class of chaperones represented by Escherichia coli Hsp31 reveals a putative catalytic triad.
|
| |
Proc Natl Acad Sci U S A,
100,
3137-3142.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Amaro,
E.Tajkhorshid,
and
Z.Luthey-Schulten
(2003).
Developing an energy landscape for the novel function of a (beta/alpha)8 barrel: ammonia conduction through HisF.
|
| |
Proc Natl Acad Sci U S A,
100,
7599-7604.
|
 |
|
|
|
|
 |
A.Douangamath,
M.Walker,
S.Beismann-Driemeyer,
M.C.Vega-Fernandez,
R.Sterner,
and
M.Wilmanns
(2002).
Structural evidence for ammonia tunneling across the (beta alpha)(8) barrel of the imidazole glycerol phosphate synthase bienzyme complex.
|
| |
Structure,
10,
185-193.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Saeed-Kothe,
and
S.G.Powers-Lee
(2002).
Specificity determining residues in ammonia- and glutamine-dependent carbamoyl phosphate synthetases.
|
| |
J Biol Chem,
277,
7231-7238.
|
 |
|
|
|
|
 |
C.T.Lemke,
and
P.L.Howell
(2002).
Substrate induced conformational changes in argininosuccinate synthetase.
|
| |
J Biol Chem,
277,
13074-13081.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Li,
T.J.Ryan,
K.J.Chave,
and
P.Van Roey
(2002).
Three-dimensional structure of human gamma -glutamyl hydrolase. A class I glatamine amidotransferase adapted for a complex substate.
|
| |
J Biol Chem,
277,
24522-24529.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.B.Thoden,
X.Huang,
F.M.Raushel,
and
H.M.Holden
(2002).
Carbamoyl-phosphate synthetase. Creation of an escape route for ammonia.
|
| |
J Biol Chem,
277,
39722-39727.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Aravind,
V.Anantharaman,
and
E.V.Koonin
(2002).
Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: implications for protein evolution in the RNA.
|
| |
Proteins,
48,
1.
|
 |
|
|
|
|
 |
M.Goto,
Y.Nakajima,
and
K.Hirotsu
(2002).
Crystal structure of argininosuccinate synthetase from Thermus thermophilus HB8. Structural basis for the catalytic action.
|
| |
J Biol Chem,
277,
15890-15896.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Korolev,
T.Skarina,
E.Evdokimova,
S.Beasley,
A.Edwards,
A.Joachimiak,
and
A.Savchenko
(2002).
Crystal structure of glutamine amidotransferase from Thermotoga maritima.
|
| |
Proteins,
49,
420-422.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Spraggon,
C.Kim,
X.Nguyen-Huu,
M.C.Yee,
C.Yanofsky,
and
S.E.Mills
(2001).
The structures of anthranilate synthase of Serratia marcescens crystallized in the presence of (i) its substrates, chorismate and glutamine, and a product, glutamate, and (ii) its end-product inhibitor, L-tryptophan.
|
| |
Proc Natl Acad Sci U S A,
98,
6021-6026.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.C.Pace,
and
C.Brenner
(2001).
The nitrilase superfamily: classification, structure and function.
|
| |
Genome Biol,
2,
REVIEWS0001.
|
 |
|
|
|
|
 |
M.M.Horvath,
and
N.V.Grishin
(2001).
The C-terminal domain of HPII catalase is a member of the type I glutamine amidotransferase superfamily.
|
| |
Proteins,
42,
230-236.
|
 |
|
|
|
|
 |
M.T.Hilgers,
and
M.L.Ludwig
(2001).
Crystal structure of the quorum-sensing protein LuxS reveals a catalytic metal site.
|
| |
Proc Natl Acad Sci U S A,
98,
11169-11174.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.V.Grishin
(2001).
KH domain: one motif, two folds.
|
| |
Nucleic Acids Res,
29,
638-643.
|
 |
|
|
|
|
 |
T.J.Klem,
Y.Chen,
and
V.J.Davisson
(2001).
Subunit interactions and glutamine utilization by Escherichia coli imidazole glycerol phosphate synthase.
|
| |
J Bacteriol,
183,
989-996.
|
 |
|
|
|
|
 |
X.Huang,
H.M.Holden,
and
F.M.Raushel
(2001).
Channeling of substrates and intermediates in enzyme-catalyzed reactions.
|
| |
Annu Rev Biochem,
70,
149-180.
|
 |
|
|
|
|
 |
A.K.Bera,
S.Chen,
J.L.Smith,
and
H.Zalkin
(2000).
Temperature-dependent function of the glutamine phosphoribosylpyrophosphate amidotransferase ammonia channel and coupling with glycinamide ribonucleotide synthetase in a hyperthermophile.
|
| |
J Bacteriol,
182,
3734-3739.
|
 |
|
|
|
|
 |
C.E.Stevenson,
F.Sargent,
G.Buchanan,
T.Palmer,
and
D.M.Lawson
(2000).
Crystal structure of the molybdenum cofactor biosynthesis protein MobA from Escherichia coli at near-atomic resolution.
|
| |
Structure,
8,
1115-1125.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.A.Denessiouk,
and
M.S.Johnson
(2000).
When fold is not important: a common structural framework for adenine and AMP binding in 12 unrelated protein families.
|
| |
Proteins,
38,
310-326.
|
 |
|
|
|
|
 |
M.A.Rishavy,
W.W.Cleland,
and
C.J.Lusty
(2000).
15N isotope effects in glutamine hydrolysis catalyzed by carbamyl phosphate synthetase: evidence for a tetrahedral intermediate in the mechanism.
|
| |
Biochemistry,
39,
7309-7315.
|
 |
|
|
|
|
 |
M.Y.Galperin,
and
N.V.Grishin
(2000).
The synthetase domains of cobalamin biosynthesis amidotransferases cobB and cobQ belong to a new family of ATP-dependent amidoligases, related to dethiobiotin synthetase.
|
| |
Proteins,
41,
238-247.
|
 |
|
|
|
|
 |
P.M.Palenchar,
C.J.Buck,
H.Cheng,
T.J.Larson,
and
E.G.Mueller
(2000).
Evidence that ThiI, an enzyme shared between thiamin and 4-thiouridine biosynthesis, may be a sulfurtransferase that proceeds through a persulfide intermediate.
|
| |
J Biol Chem,
275,
8283-8286.
|
 |
|
|
|
|
 |
X.Du,
I.G.Choi,
R.Kim,
W.Wang,
J.Jancarik,
H.Yokota,
and
S.H.Kim
(2000).
Crystal structure of an intracellular protease from Pyrococcus horikoshii at 2-A resolution.
|
| |
Proc Natl Acad Sci U S A,
97,
14079-14084.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.Zhang,
L.Rychlewski,
K.Pawłowski,
J.S.Fetrow,
J.Skolnick,
and
A.Godzik
(1999).
From fold predictions to function predictions: automation of functional site conservation analysis for functional genome predictions.
|
| |
Protein Sci,
8,
1104-1115.
|
 |
|
|
|
|
 |
C.H.Weber,
Y.S.Park,
S.Sanker,
C.Kent,
and
M.L.Ludwig
(1999).
A prototypical cytidylyltransferase: CTP:glycerol-3-phosphate cytidylyltransferase from bacillus subtilis.
|
| |
Structure,
7,
1113-1124.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.G.Mueller,
and
P.M.Palenchar
(1999).
Using genomic information to investigate the function of ThiI, an enzyme shared between thiamin and 4-thiouridine biosynthesis.
|
| |
Protein Sci,
8,
2424-2427.
|
 |
|
|
|
|
 |
F.M.Raushel,
J.B.Thoden,
and
H.M.Holden
(1999).
The amidotransferase family of enzymes: molecular machines for the production and delivery of ammonia.
|
| |
Biochemistry,
38,
7891-7899.
|
 |
|
|
|
|
 |
J.B.Thoden,
X.Huang,
F.M.Raushel,
and
H.M.Holden
(1999).
The small subunit of carbamoyl phosphate synthetase: snapshots along the reaction pathway.
|
| |
Biochemistry,
38,
16158-16166.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Knöchel,
A.Ivens,
G.Hester,
A.Gonzalez,
R.Bauerle,
M.Wilmanns,
K.Kirschner,
and
J.N.Jansonius
(1999).
The crystal structure of anthranilate synthase from Sulfolobus solfataricus: functional implications.
|
| |
Proc Natl Acad Sci U S A,
96,
9479-9484.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.M.Larsen,
S.K.Boehlein,
S.M.Schuster,
N.G.Richards,
J.B.Thoden,
H.M.Holden,
and
I.Rayment
(1999).
Three-dimensional structure of Escherichia coli asparagine synthetase B: a short journey from substrate to product.
|
| |
Biochemistry,
38,
16146-16157.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Hewagama,
H.I.Guy,
M.Chaparian,
and
D.R.Evans
(1998).
The function of Glu338 in the catalytic triad of the carbamoyl phosphate synthetase amidotransferase domain.
|
| |
Biochim Biophys Acta,
1388,
489-499.
|
 |
|
|
|
|
 |
F.M.Raushel,
J.B.Thoden,
G.D.Reinhart,
and
H.M.Holden
(1998).
Carbamoyl phosphate synthetase: a crooked path from substrates to products.
|
| |
Curr Opin Chem Biol,
2,
624-632.
|
 |
|
|
|
|
 |
H.M.Holden,
J.B.Thoden,
and
F.M.Raushel
(1998).
Carbamoyl phosphate synthetase: a tunnel runs through it.
|
| |
Curr Opin Struct Biol,
8,
679-685.
|
 |
|
|
|
|
 |
J.B.Thoden,
S.G.Miran,
J.C.Phillips,
A.J.Howard,
F.M.Raushel,
and
H.M.Holden
(1998).
Carbamoyl phosphate synthetase: caught in the act of glutamine hydrolysis.
|
| |
Biochemistry,
37,
8825-8831.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.L.Smith
(1998).
Glutamine PRPP amidotransferase: snapshots of an enzyme in action.
|
| |
Curr Opin Struct Biol,
8,
686-694.
|
 |
|
|
|
|
 |
J.L.Smith,
and
A.Thompson
(1998).
Reactivity of selenomethionine--dents in the magic bullet?
|
| |
Structure,
6,
815-819.
|
 |
|
|
|
|
 |
M.Rizzi,
M.Bolognesi,
and
A.Coda
(1998).
A novel deamido-NAD+-binding site revealed by the trapped NAD-adenylate intermediate in the NAD+ synthetase structure.
|
| |
Structure,
6,
1129-1140.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Cantoni,
M.Branzoni,
M.Labò,
M.Rizzi,
and
G.Riccardi
(1998).
The MTCY428.08 gene of Mycobacterium tuberculosis codes for NAD+ synthetase.
|
| |
J Bacteriol,
180,
3218-3221.
|
 |
|
|
|
|
 |
W.M.Liu,
and
K.C.Chou
(1998).
Singular points of protein beta-sheets.
|
| |
Protein Sci,
7,
2324-2330.
|
 |
|
|
|
|
 |
H.Savage,
G.Montoya,
C.Svensson,
J.D.Schwenn,
and
I.Sinning
(1997).
Crystal structure of phosphoadenylyl sulphate (PAPS) reductase: a new family of adenine nucleotide alpha hydrolases.
|
| |
Structure,
5,
895-906.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.B.Thoden,
H.M.Holden,
G.Wesenberg,
F.M.Raushel,
and
I.Rayment
(1997).
Structure of carbamoyl phosphate synthetase: a journey of 96 A from substrate to product.
|
| |
Biochemistry,
36,
6305-6316.
|
 |
|
|
|
|
 |
M.O'Gara,
G.M.Adams,
W.Gong,
R.Kobayashi,
R.M.Blumenthal,
and
X.Cheng
(1997).
Expression, purification, mass spectrometry, crystallization and multiwavelength anomalous diffraction of selenomethionyl PvuII DNA methyltransferase (cytosine-N4-specific).
|
| |
Eur J Biochem,
247,
1009-1018.
|
 |
|
|
|
|
 |
B.Xiang,
and
G.D.Markham
(1996).
The conformation of inosine 5'-monophosphate (IMP) bound to IMP dehydrogenase determined by transferred nuclear overhauser effect spectroscopy.
|
| |
J Biol Chem,
271,
27531-27535.
|
 |
|
|
|
|
 |
J.H.Kim,
J.M.Krahn,
D.R.Tomchick,
J.L.Smith,
and
H.Zalkin
(1996).
Structure and function of the glutamine phosphoribosylpyrophosphate amidotransferase glutamine site and communication with the phosphoribosylpyrophosphate site.
|
| |
J Biol Chem,
271,
15549-15557.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.N.Isupov,
G.Obmolova,
S.Butterworth,
M.A.Badet-Denisot,
B.Badet,
I.Polikarpov,
J.A.Littlechild,
and
A.Teplyakov
(1996).
Substrate binding is required for assembly of the active conformation of the catalytic site in Ntn amidotransferases: evidence from the 1.8 A crystal structure of the glutaminase domain of glucosamine 6-phosphate synthase.
|
| |
Structure,
4,
801-810.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Rizzi,
C.Nessi,
A.Mattevi,
A.Coda,
M.Bolognesi,
and
A.Galizzi
(1996).
Crystal structure of NH3-dependent NAD+ synthetase from Bacillus subtilis.
|
| |
EMBO J,
15,
5125-5134.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Rizzi,
C.Nessi,
M.Bolognesi,
A.Coda,
and
A.Galizzi
(1996).
Crystallization of NAD+ synthetase from Bacillus subtilis.
|
| |
Proteins,
26,
236-238.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

