 |
PDBsum entry 1vcm

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.4.2
- Ctp synthase (glutamine hydrolyzing).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
UTP + L-glutamine + ATP + H2O = CTP + L-glutamate + ADP + phosphate + 2 H+
|
 |
 |
 |
 |
 |
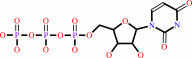 UTP
UTP
|
+
|
 L-glutamine
L-glutamine
|
+
|
 ATP
ATP
|
+
|
H2O
|
=
|
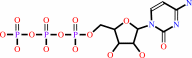 CTP
CTP
|
+
|
 L-glutamate
L-glutamate
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
12:1413-1423
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of CTP synthetase reveal ATP, UTP, and glutamine binding sites.
|
|
M.Goto,
R.Omi,
N.Nakagawa,
I.Miyahara,
K.Hirotsu.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
CTP synthetase (CTPs) catalyzes the last step in CTP biosynthesis, in which
ammonia generated at the glutaminase domain reacts with the ATP-phosphorylated
UTP at the synthetase domain to give CTP. Glutamine hydrolysis is active in the
presence of ATP and UTP and is stimulated by the addition of GTP. We report the
crystal structures of Thermus thermophilus HB8 CTPs alone, CTPs with 3SO4(2-),
and CTPs with glutamine. The enzyme is folded into a homotetramer with a
cross-shaped structure. Based on the binding mode of sulfate anions to the
synthetase site, ATP and UTP are computer modeled into CTPs with a geometry
favorable for the reaction. Glutamine bound to the glutaminase domain is
situated next to the triad of Glu-His-Cys as a catalyst and a water molecule.
Structural information provides an insight into the conformational changes
associated with the binding of ATP and UTP and the formation of the GTP binding
site.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 8.
Figure 8. Putative Conformational Change upon Binding of
ATP and UTPThe glutaminase domain (brown) is rotated toward the
synthetase domain (blue) on the graphics to form a computer
model of the compact molecule in the closed form. The consensus
sequence (green) specific for GTP on the glutaminase domain
approaches that (green) on the synthetase domain to form a
binding site for GTP. ATP and UTP modeled into the closed form
are drawn in red.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2004,
12,
1413-1423)
copyright 2004.
|
|
| |
Figure was
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.L.Liu
(2011).
The enigmatic cytoophidium: Compartmentation of CTP synthase via filament formation.
|
| |
Bioessays,
33,
159-164.
|
 |
|
|
|
|
 |
F.A.Lunn,
J.E.Macdonnell,
and
S.L.Bearne
(2008).
Structural Requirements for the Activation of Escherichia coli CTP Synthase by the Allosteric Effector GTP Are Stringent, but Requirements for Inhibition Are Lax.
|
| |
J Biol Chem,
283,
2010-2020.
|
 |
|
|
|
|
 |
M.Morar,
A.A.Hoskins,
J.Stubbe,
and
S.E.Ealick
(2008).
Formylglycinamide ribonucleotide amidotransferase from Thermotoga maritima: structural insights into complex formation.
|
| |
Biochemistry,
47,
7816-7830.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.F.Chang,
and
G.M.Carman
(2008).
CTP synthetase and its role in phospholipid synthesis in the yeast Saccharomyces cerevisiae.
|
| |
Prog Lipid Res,
47,
333-339.
|
 |
|
|
|
|
 |
A.Fijolek,
A.Hofer,
and
L.Thelander
(2007).
Expression, purification, characterization, and in vivo targeting of trypanosome CTP synthetase for treatment of African sleeping sickness.
|
| |
J Biol Chem,
282,
11858-11865.
|
 |
|
|
|
|
 |
M.G.Choi,
and
G.M.Carman
(2007).
Phosphorylation of human CTP synthetase 1 by protein kinase A: identification of Thr455 as a major site of phosphorylation.
|
| |
J Biol Chem,
282,
5367-5377.
|
 |
|
|
|
|
 |
M.J.Higgins,
P.R.Graves,
and
L.M.Graves
(2007).
Regulation of human cytidine triphosphate synthetase 1 by glycogen synthase kinase 3.
|
| |
J Biol Chem,
282,
29493-29503.
|
 |
|
|
|
|
 |
O.Braun,
M.Knipp,
S.Chesnov,
and
M.Vasák
(2007).
Specific reactions of S-nitrosothiols with cysteine hydrolases: A comparative study between dimethylargininase-1 and CTP synthetase.
|
| |
Protein Sci,
16,
1522-1534.
|
 |
|
|
|
|
 |
S.Mouilleron,
and
B.Golinelli-Pimpaneau
(2007).
Conformational changes in ammonia-channeling glutamine amidotransferases.
|
| |
Curr Opin Struct Biol,
17,
653-664.
|
 |
|
|
|
|
 |
Y.F.Chang,
S.S.Martin,
E.P.Baldwin,
and
G.M.Carman
(2007).
Phosphorylation of human CTP synthetase 1 by protein kinase C: identification of Ser(462) and Thr(455) as major sites of phosphorylation.
|
| |
J Biol Chem,
282,
17613-17622.
|
 |
|
|
|
|
 |
P.Kursula,
S.Flodin,
M.Ehn,
M.Hammarström,
H.Schüler,
P.Nordlund,
and
P.Stenmark
(2006).
Structure of the synthetase domain of human CTP synthetase, a target for anticancer therapy.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
613-617.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.S.Han,
A.Sreenivas,
M.G.Choi,
Y.F.Chang,
S.S.Martin,
E.P.Baldwin,
and
G.M.Carman
(2005).
Expression of Human CTP synthetase in Saccharomyces cerevisiae reveals phosphorylation by protein kinase A.
|
| |
J Biol Chem,
280,
38328-38336.
|
 |
|
|
|
|
 |
J.A.Endrizzi,
H.Kim,
P.M.Anderson,
and
E.P.Baldwin
(2005).
Mechanisms of product feedback regulation and drug resistance in cytidine triphosphate synthetases from the structure of a CTP-inhibited complex.
|
| |
Biochemistry,
44,
13491-13499.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Willemoës,
A.Mølgaard,
E.Johansson,
and
J.Martinussen
(2005).
Lid L11 of the glutamine amidotransferase domain of CTP synthase mediates allosteric GTP activation of glutaminase activity.
|
| |
FEBS J,
272,
856-864.
|
 |
|
|
|
|
 |
W.L.Yang,
and
G.M.Carman
(1995).
Phosphorylation of CTP synthetase from Saccharomyces cerevisiae by protein kinase C.
|
| |
J Biol Chem,
270,
14983-14988.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

