Do data resources managed by EMBL-EBI and our collaborators make a difference to your work? Please take 10 minutes to fill in our annual user survey, and help us make the case for why sustaining open data resources is critical for life sciences research. We appreciate your responses prior to closing of the survey on Monday 17th June.
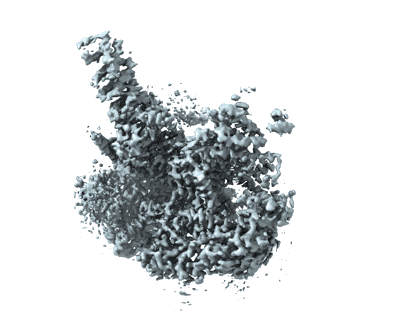
State NE1 nucleolar 60S ribosome biogenesis intermediate - Overall map
Resolution: 2.76 Å
EM Method: Single-particle
Fitted PDBs: 7u0h
Q-score: 0.592
Cruz VE, Sekulski K, Peddada N, Sailer C, Balasubramanian S, Weirich CS, Stengel F, Erzberger JP
Nat Struct Mol Biol (2022) 29 pp. 1228-1238 [ Pubmed: 36482249 DOI: doi:10.1038/s41594-022-00874-9 ]
- Its2 rrna (27 kDa, RNA from Saccharomyces cerevisiae BY4741)
- Nucleolar gtp-binding protein 2 (1 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l38 (8 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar gtp-binding protein 1 (74 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l23-a (14 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Eukaryotic translation initiation factor 6 (26 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l9-a (21 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l34-a (13 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l14-a (15 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l17-a (20 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l33-a (12 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l30 (11 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l31-a (12 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein nop53 (52 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l22-a (13 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l37-a (9 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l43-a (10 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein nsa2 (29 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l16-a (22 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l13-a (22 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l7-a (27 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein 15 (25 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Upf0642 protein ybl028c (12 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l27-a (15 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l36-a (11 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome assembly factor mrt4 (27 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l25 (15 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l6-a (20 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 27s pre-rrna (guanosine(2922)-2'-o)-methyltransferase (96 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l28 (16 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l21-a (18 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 25s rrna (1 MDa, RNA from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l35-a (13 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l3 (43 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l2-a (27 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l26-a (14 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein rlp24 (24 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 5.8s rrna (50 kDa, RNA from Saccharomyces cerevisiae BY4741)
- Early nucleoplasmic 60s intermediate purified with tags on nop53 and spb1. (2 MDa, Complex from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l32 (14 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Zinc ion (65 Da, Ligand)
- Ribosome biogenesis protein rlp7 (36 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l19-a (21 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l4-a (39 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nuclear gtp-binding protein nug1 (57 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l8-a (28 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l20-a (20 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l15-a (24 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Magnesium ion (24 Da, Ligand)
- Pescadillo homolog (69 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Proteasome-interacting protein cic1 (42 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l18-a (20 kDa, Protein from Saccharomyces cerevisiae BY4741)
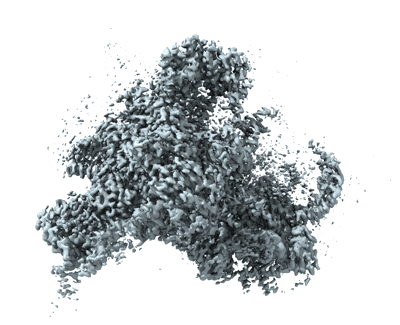
State E2 nucleolar 60S ribosomal biogenesis intermediate - Overall map
Resolution: 3.04 Å
EM Method: Single-particle
Fitted PDBs: 7nac
Q-score: 0.549
Cruz VE, Sekulski K, Peddada N, Sailer C, Balasubramanian S, Weirich CS, Stengel F, Erzberger JP
Nat Struct Mol Biol (2022) 29 pp. 1228-1238 [ DOI: doi:10.1038/s41594-022-00874-9 Pubmed: 36482249 ]
- Nuclear gtp-binding protein nug1 (57 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein nsa2 (29 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l32 (14 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Atp-dependent rna helicase has1 (56 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosome subunit biogenesis protein nip7 (20 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein rlp7 (36 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein rlp24 (24 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l4-a (39 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l26-a (14 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Rrna-processing protein ebp2 (49 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l14-a (15 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein 15 (25 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l37-a (9 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein erb1 (91 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l7-a (27 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l22-a (13 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l33-a (12 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l30 (11 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Noc2 isoform 1 (81 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l9-a (21 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l19-a (21 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Atp-dependent rrna helicase spb4 (69 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosomal protein (24 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Proteasome-interacting protein cic1 (42 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 5.8s ribosomal rna gene and internal transcribed spacer 2 (27 kDa, RNA from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l38 (8 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l13-a (22 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l25 (15 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l3 (43 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Pescadillo homolog (69 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l35-a (13 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein brx1 (33 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l31-a (12 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar complex-associated protein 3 (75 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l8-a (28 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Eukaryotic translation initiation factor 6 (26 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 25s rrna (cytosine(2870)-c(5))-methyltransferase (69 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l27-a (15 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Rrp17 isoform 1 (28 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar 60s intermediate purified with tags on ytm1 and spb4. (3 MDa, Complex from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l15-a (24 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l34-a (13 kDa, Protein from Saccharomyces cerevisiae BY4741)
- S-adenosyl-l-homocysteine (384 Da, Ligand)
- 60s ribosomal protein l21-a (18 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 25s rrna (1 MDa, RNA from Saccharomyces cerevisiae BY4741)
- 27s pre-rrna (guanosine(2922)-2'-o)-methyltransferase (96 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l18-a (20 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ytm1 isoform 1 (51 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l36-a (11 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Zinc ion (65 Da, Ligand)
- Nucleolar gtp-binding protein 1 (74 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Upf0642 protein ybl028c (12 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 5.8s rrna (50 kDa, RNA from Saccharomyces cerevisiae BY4741)
- 60s ribosomal subunit assembly/export protein loc1 (23 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome assembly factor mrt4 (27 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l20-a (20 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l17-a (20 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l16-a (22 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l6-a (20 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar protein 16 (26 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l23-a (14 kDa, Protein from Saccharomyces cerevisiae BY4741)
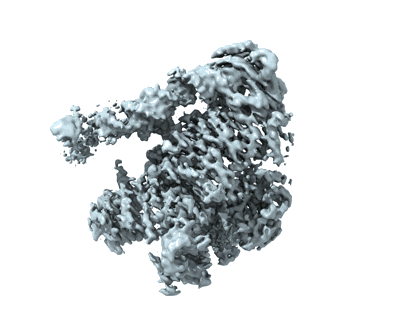
State E2 nucleolar 60S ribosomal biogenesis intermediate - Spb4 local refinement model
Resolution: 3.04 Å
EM Method: Single-particle
Fitted PDBs: 7nad
Q-score: 0.547
Cruz VE, Sekulski K, Peddada N, Sailer C, Balasubramanian S, Weirich CS, Stengel F, Erzberger JP
Nat Struct Mol Biol (2022) 29 pp. 1228-1238 [ DOI: doi:10.1038/s41594-022-00874-9 Pubmed: 36482249 ]
- Ribosome biogenesis protein rlp24 (24 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 25s rrna (225 kDa, RNA from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l37-a (9 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l22-a (13 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l23-a (4 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l30 (11 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Noc2 isoform 1 (81 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Rpl38 isoform 1 (8 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l19-a (21 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Pescadillo homolog (12 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l25 (15 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l3 (43 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l31-a (12 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 5.8s rrna (23 kDa, RNA from Saccharomyces cerevisiae BY4741)
- Rrp17 isoform 1 (28 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l27-a (15 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Spb1 isoform 1 (96 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar 60s intermediate purified with tags on ytm1 and spb4. (3 MDa, Complex from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l34-a (13 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Rlp7 isoform 1 (36 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ytm1 isoform 1 (51 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Rpl8a isoform 1 (28 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein erb1 (48 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Spb4 isoform 1 (69 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Zinc ion (65 Da, Ligand)
- 60s ribosomal protein l17-a (20 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Noc3 isoform 1 (75 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar gtp-binding protein 1 (74 kDa, Protein from Saccharomyces cerevisiae BY4741)
State E2 nucleolar 60S ribosomal biogenesis intermediate - Spb1-MTD locally refined map
Resolution: 3.13 Å
EM Method: Single-particle
Fitted PDBs: 7naf
Q-score: 0.513
Cruz VE, Sekulski K, Peddada N, Sailer C, Balasubramanian S, Weirich CS, Stengel F, Erzberger JP
Nat Struct Mol Biol (2022) 29 pp. 1228-1238 [ DOI: doi:10.1038/s41594-022-00874-9 Pubmed: 36482249 ]
- 25s rrna (cytosine(2870)-c(5))-methyltransferase (3 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Water (18 Da, Ligand)
- 60s ribosomal protein l30 (4 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosomal rna-processing protein 17 (13 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l3 (42 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l31-a (7 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar complex protein 2 (4 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 27s pre-rrna (guanosine(2922)-2'-o)-methyltransferase (45 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein rlp24 (7 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nuclear gtp-binding protein nug1 (977 Da, Protein from Saccharomyces cerevisiae BY4741)
- Eukaryotic translation initiation factor 6 (26 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l22-a (2 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 25s rrna (121 kDa, RNA from Saccharomyces cerevisiae BY4741)
- Nucleolar 60s intermediate purified with tags on ytm1 and spb4. (3 MDa, Complex from Saccharomyces cerevisiae BY4741)
- 60s ribosome ribosomal protein l19a (7 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Upf0642 protein ybl028c (646 Da, Protein from Saccharomyces cerevisiae BY4741)
- S-adenosyl-l-homocysteine (384 Da, Ligand)
- 60s ribosome biogenesis factor nog1 (8 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l23-a (14 kDa, Protein from Saccharomyces cerevisiae BY4741)
State E2 nucleolar 60S ribosomal intermediate - Local Map for Noc2/Noc3 region
Resolution: 3.17 Å
EM Method: Single-particle
Fitted PDBs: 7r6k
Q-score: 0.495
Cruz VE, Sekulski K, Peddada N, Sailer C, Balasubramanian S, Weirich CS, Stengel F, Erzberger JP
Nat Struct Mol Biol (2022) 29 pp. 1228-1238 [ DOI: doi:10.1038/s41594-022-00874-9 Pubmed: 36482249 ]
- Ribosome biogenesis protein nsa2 (29 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein erb1 (91 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 25s rrna (49 kDa, RNA from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein brx1 (33 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosome subunit biogenesis protein nip7 (20 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 27s pre-rrna (guanosine(2922)-2'-o)-methyltransferase (96 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Rrp17 isoform 1 (1 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar 60s intermediate purified with tags on ytm1 and spb4 (3 MDa, Complex from Saccharomyces cerevisiae BY4741)
- Nucleolar complex-associated protein 3 (75 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l8-a (28 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal subunit assembly/export protein loc1 (23 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Rrna-processing protein ebp2 (49 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 25s rrna (cytosine(2870)-c(5))-methyltransferase (69 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l36-a (11 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l34-a (13 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l15-a (24 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar complex protein 2 (81 kDa, Protein from Saccharomyces cerevisiae BY4741)
State E2 nucleolar 60S ribosome biogenesis intermediate - Foot region map
Resolution: 2.98 Å
EM Method: Single-particle
Fitted PDBs: 7r6q
Q-score: 0.576
Cruz VE, Sekulski K, Peddada N, Sailer C, Balasubramanian S, Weirich CS, Stengel F, Erzberger JP
Nat Struct Mol Biol (2022) 29 pp. 1228-1238 [ DOI: doi:10.1038/s41594-022-00874-9 Pubmed: 36482249 ]
- Atp-dependent rna helicase has1 (56 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein rlp7 (36 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar 60s intermediate purified with tags on ytm1 and spb4 (3 MDa, Complex from Saccharomyces cerevisiae BY4741)
- 25s rrna (95 kDa, RNA from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein 15 (25 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Proteasome-interacting protein cic1 (42 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosome biogenesis factor spb1 (8 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l34-a (4 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l25 (15 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l13-a (22 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Pescadillo homolog (69 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Its-2 (27 kDa, RNA from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l8-a (28 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 5.8s rrna (18 kDa, RNA from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l15-a (24 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l35 (2 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein erb1 (91 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l27 (1 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l36-a (11 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar protein 16 (26 kDa, Protein from Saccharomyces cerevisiae BY4741)
State E1 nucleolar 60S ribosome biogenesis intermediate - Spb4 locally refined map
Resolution: 3.07 Å
EM Method: Single-particle
Fitted PDBs: 7r72
Q-score: 0.519
Cruz VE, Sekulski K, Peddada N, Sailer C, Balasubramanian S, Weirich CS, Stengel F, Erzberger JP
Nat Struct Mol Biol (2022) 29 pp. 1228-1238 [ DOI: doi:10.1038/s41594-022-00874-9 Pubmed: 36482249 ]
- Ribosome biogenesis protein rlp7 (36 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l37-a (9 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l22-a (13 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar complex protein 2 (81 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l30 (11 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Pescadillo homolog (12 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Atp-dependent rrna helicase spb4 (69 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l19-a (21 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l38 (8 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l25 (15 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l3 (43 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein ytm1 (51 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l31-a (12 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar complex-associated protein 3 (75 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 25s rrna (207 kDa, RNA from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l8-a (28 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 5.8s rrna (23 kDa, RNA from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l27-a (15 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l34-a (13 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar 60s intermediate purified with tags on ytm1 and spb4. (3 MDa, Complex from Saccharomyces cerevisiae BY4741)
- 27s pre-rrna (guanosine(2922)-2'-o)-methyltransferase (96 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein erb1 (48 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Zinc ion (65 Da, Ligand)
- 60s ribosomal protein l17-a (20 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein rlp24 (24 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar gtp-binding protein 1 (74 kDa, Protein from Saccharomyces cerevisiae BY4741)
State E1 nucleolar 60S ribosome biogenesis intermediate - Composite model
Resolution: 3.04 Å
EM Method: Single-particle
Fitted PDBs: 7r7a
Q-score: 0.527
Cruz VE, Sekulski K, Peddada N, Sailer C, Balasubramanian S, Weirich CS, Stengel F, Erzberger JP
Nat Struct Mol Biol (2022) 29 pp. 1228-1238 [ DOI: doi:10.1038/s41594-022-00874-9 Pubmed: 36482249 ]
- Nuclear gtp-binding protein nug1 (57 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein nsa2 (29 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l32 (14 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Atp-dependent rna helicase has1 (56 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosome subunit biogenesis protein nip7 (20 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein rlp7 (36 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein rlp24 (24 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l4-a (39 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l26-a (14 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Rrna-processing protein ebp2 (49 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l14-a (15 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein 15 (25 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l37-a (9 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein erb1 (91 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l7-a (27 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar complex protein 2 (81 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l22-a (13 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l33-a (12 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l30 (11 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l9-a (21 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l19-a (21 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Atp-dependent rrna helicase spb4 (69 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosomal protein (24 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Proteasome-interacting protein cic1 (42 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 5.8s ribosomal rna gene and internal transcribed spacer 2 (27 kDa, RNA from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l38 (8 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l13-a (22 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l25 (15 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l3 (43 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Pescadillo homolog (69 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 25s rrna (1 MDa, RNA from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l35-a (13 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein brx1 (33 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l6-a (19 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l31-a (12 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar complex-associated protein 3 (75 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l8-a (28 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Eukaryotic translation initiation factor 6 (26 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 25s rrna (cytosine(2870)-c(5))-methyltransferase (69 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l27-a (15 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar 60s intermediate purified with tags on ytm1 and spb4. (3 MDa, Complex from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l15-a (24 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l34-a (13 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l21-a (18 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 27s pre-rrna (guanosine(2922)-2'-o)-methyltransferase (96 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l18-a (20 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ytm1 isoform 1 (51 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l36-a (11 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Zinc ion (65 Da, Ligand)
- Upf0642 protein ybl028c (12 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 5.8s rrna (50 kDa, RNA from Saccharomyces cerevisiae BY4741)
- 60s ribosomal subunit assembly/export protein loc1 (23 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome assembly factor mrt4 (27 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l20-a (20 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l17-a (20 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l16-a (22 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar gtp-binding protein 1 (74 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar protein 16 (26 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l23-a (14 kDa, Protein from Saccharomyces cerevisiae BY4741)
State E2 nucleolar 60S ribosomal biogenesis intermediate - L1 stalk local map
Resolution: 3.71 Å
EM Method: Single-particle
Fitted PDBs: 7r7c
Q-score: 0.401
Cruz VE, Sekulski K, Peddada N, Sailer C, Balasubramanian S, Weirich CS, Stengel F, Erzberger JP
Nat Struct Mol Biol (2022) 29 pp. 1228-1238 [ DOI: doi:10.1038/s41594-022-00874-9 Pubmed: 36482249 ]
- Rrna-processing protein ebp2 (11 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 27s pre-rrna (guanosine(2922)-2'-o)-methyltransferase (1 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosomal protein l1-a (24 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 60s ribosome subunit biogenesis protein nip7 (20 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein brx1 (33 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein erb1 (6 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar gtp-binding protein 1 (6 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Nucleolar 60s intermediate purified with tags on ytm1 and spb4 (3 MDa, Complex from Saccharomyces cerevisiae BY4741)
- 60s ribosomal subunit assembly/export protein loc1 (23 kDa, Protein from Saccharomyces cerevisiae BY4741)
- 25s rrna (72 kDa, RNA from Saccharomyces cerevisiae BY4741)
- 25s rrna (cytosine(2870)-c(5))-methyltransferase (69 kDa, Protein from Saccharomyces cerevisiae BY4741)
- Ribosome biogenesis protein nsa2 homolog (11 kDa, Protein from Saccharomyces cerevisiae BY4741)
Structure of pre-60S particle bound to DRG1(AFG2)
Prattes M, Grishkovskaya I, Hodirnau VV, Hetzmannseder C, Zisser G, Sailer C, Kargas V, Loibl M, Gerhalter M, Kofler L, Warren AJ, Stengel F, Haselbach D, Bergler H
Nat Struct Mol Biol (2022) [ Pubmed: 36097293 DOI: 10.1038/s41594-022-00832-5 ]
Image sets: