Nitrile hydratase
Iron-dependent nitrile hydratases (NHases) are mononuclear iron enzymes that catalyse the hydration of a large number of diverse nitriles to their corresponding amides. NHases are activated through the photochemical activation of water in which a bound NO is replaced with HO-. Organisms expressing NHases are capable of utilising aliphatic nitriles as the sole source of nitrogen.
NHases consist of alpha and beta heterodimers. These enzymes have importance as biocatalysts, for example in the production of acrylamide, and in bioremediation and have been efficiently used for the industrial production of acrylamide from acrylonitrile and for the removal of nitriles from wastewater.
Reference Protein and Structure
- Sequences
-
P13448
 (4.2.1.84)
(4.2.1.84)
P13449 (4.2.1.84)
(4.2.1.84)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Rhodococcus erythropolis (Bacteria)

- PDB
-
2ahj
- NITRILE HYDRATASE COMPLEXED WITH NITRIC OXIDE
(1.7 Å)



- Catalytic CATH Domains
-
3.90.330.10
 1.10.472.20
1.10.472.20  (see all for 2ahj)
(see all for 2ahj)
- Cofactors
- Iron(3+) (1), Water (1) Metal MACiE
Enzyme Reaction (EC:4.2.1.84)
Enzyme Mechanism
Introduction
In this mechanism proposal, Cys-SO- acts as the nucleophile, performing a direct nucleophilic attack on the metal-coordinated nitrile. The resulting cyclic intermediate is subsequently cleaved through attack of the axial cysteine on the sulfenate, thereby forming a disulfide bond. In this mechanism, nitrile hydration occurs without directly involving a water molecule. Subsequent water-mediated disulfide cleavage regenerates the active site.
Catalytic Residues Roles
| UniProt | PDB* (2ahj) | ||
| Cys110 | Cys109A | Acts as a catalytic nucleophile, forms the disulfide bond "switch". | nucleophile, nucleofuge, metal ligand |
| Cys115 (ptm) | Cso114A (ptm) | Acts as the initial catalytic nucleophile. | nucleophile, metal ligand, proton acceptor, proton donor, electrofuge |
| Arg56 | Arg56B | Helps stabilise the reactive intermediates, and maintain the correct protonation state of the active site. | proton acceptor, electrostatic stabiliser, proton donor |
| Cys113 (ptm), Ser114 (main-N) | Csd112A (ptm), Ser113A (main-N) | Forms part of the iron binding site. | metal ligand |
| Ser114, Tyr72 | Ser113A, Tyr72B | Forms part of the Ser-Tyr-Tyr catalytic triad, acts as a general acid/base. | proton relay, proton acceptor, proton donor |
| Tyr76 | Tyr76B | Forms part of the Ser-Tyr-Tyr catalytic triad, responsible for activating Tyr72 and Ser113 as general acid/bases. | increase basicity, increase acidity |
Chemical Components
bimolecular nucleophilic addition, proton transfer, bimolecular nucleophilic substitution, tautomerisation (not keto-enol), native state of enzyme regeneratedReferences
- Hopmann KH (2014), Inorg Chem, 53, 2760-2762. Full Reaction Mechanism of Nitrile Hydratase: A Cyclic Intermediate and an Unexpected Disulfide Switch. DOI:10.1021/ic500091k. PMID:24597943.
- Wu Y et al. (2017), Eur J Inorg Chem, 2017, 840-843. C=N Bond Activation and Hydration by an Iron(III) Complex with Asymmetric Sulfur Oxygenation. DOI:10.1002/ejic.201601565.
- Kayanuma M et al. (2016), J Phys Chem B, 120, 3259-3266. Catalytic Mechanism of Nitrile Hydratase Subsequent to Cyclic Intermediate Formation: A QM/MM Study. DOI:10.1021/acs.jpcb.5b11363. PMID:27007978.
- Light KM et al. (2015), Chem Sci, 6, 6280-6294. Spectroscopic and computational studies of nitrile hydratase: insights into geometric and electronic structure and the mechanism of amide synthesis. DOI:10.1039/c5sc02012c. PMID:26508996.
- Yamanaka Y et al. (2015), Angew Chem Int Ed Engl, 54, 10763-10767. Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile. DOI:10.1002/anie.201502731. PMID:26333053.
- MacDonald CA et al. (2015), Comput Theor Chem, 1070, 48-54. Competing nitrile hydratase catalytic mechanisms: Is cysteine-sulfenic acid acting as a nucleophile? DOI:10.1016/j.comptc.2015.07.010.
- Martinez S et al. (2015), J Biol Inorg Chem, 20, 885-894. Analyzing the catalytic role of active site residues in the Fe-type nitrile hydratase from Comamonas testosteroni Ni1. DOI:10.1007/s00775-015-1273-3. PMID:26077812.
- Kayanuma M et al. (2015), Chem Phys Lett, 623, 8-13. A QM/MM study of the initial steps of catalytic mechanism of nitrile hydratase. DOI:10.1016/j.cplett.2015.01.039.
- Martinez S et al. (2014), J Am Chem Soc, 136, 1186-1189. The Active Site Sulfenic Acid Ligand in Nitrile Hydratases Can Function as a Nucleophile. DOI:10.1021/ja410462j. PMID:24383915.
- Gumataotao N et al. (2013), J Biol Chem, 288, 15532-15536. Identification of an Active Site-bound Nitrile Hydratase Intermediate through Single Turnover Stopped-flow Spectroscopy. DOI:10.1074/jbc.m112.398909. PMID:23589282.
- Yamanaka Y et al. (2010), J Biol Inorg Chem, 15, 655-665. Kinetic and structural studies on roles of the serine ligand and a strictly conserved tyrosine residue in nitrile hydratase. DOI:10.1007/s00775-010-0632-3. PMID:20221653.
- Mitra S et al. (2007), J Biol Chem, 282, 7397-7404. Unraveling the Catalytic Mechanism of Nitrile Hydratases. DOI:10.1074/jbc.m604117200. PMID:17150969.
- Endo I et al. (2001), J Inorg Biochem, 83, 247-253. Fe-type nitrile hydratase. DOI:10.1016/s0162-0134(00)00171-9. PMID:11293544.
- Kobayashi M et al. (2000), Curr Opin Chem Biol, 4, 531-547. Nitrile Hydrolases. DOI:10.1007/1-4020-5377-0_30. PMID:10679370.
- Piersma SR et al. (2000), J Inorg Biochem, 80, 283-288. Arginine 56 mutation in the β subunit of nitrile hydratase: importance of hydrogen bonding to the non-heme iron center. DOI:10.1016/s0162-0134(00)00076-3. PMID:11001100.
- Murakami T et al. (2000), Protein Sci, 9, 1024-1030. Post-translational modification is essential for catalytic activity of nitrile hydratase. DOI:10.1110/ps.9.5.1024. PMID:10850812.
- Endo I et al. (1999), Trends Biotechnol, 17, 244-248. An enzyme controlled by light: the molecular mechanism of photoreactivity in nitrile hydratase. DOI:10.1016/s0167-7799(99)01303-7. PMID:10354562.
- Nagashima S et al. (1998), Nat Struct Biol, 5, 347-351. Novel non-heme iron center of nitrile hydratase with a claw setting of oxygen atoms. DOI:10.1038/nsb0598-347. PMID:9586994.
- Huang W et al. (1997), Structure, 5, 691-699. Crystal structure of nitrile hydratase reveals a novel iron centre in a novel fold. DOI:10.1016/s0969-2126(97)00223-2. PMID:9195885.
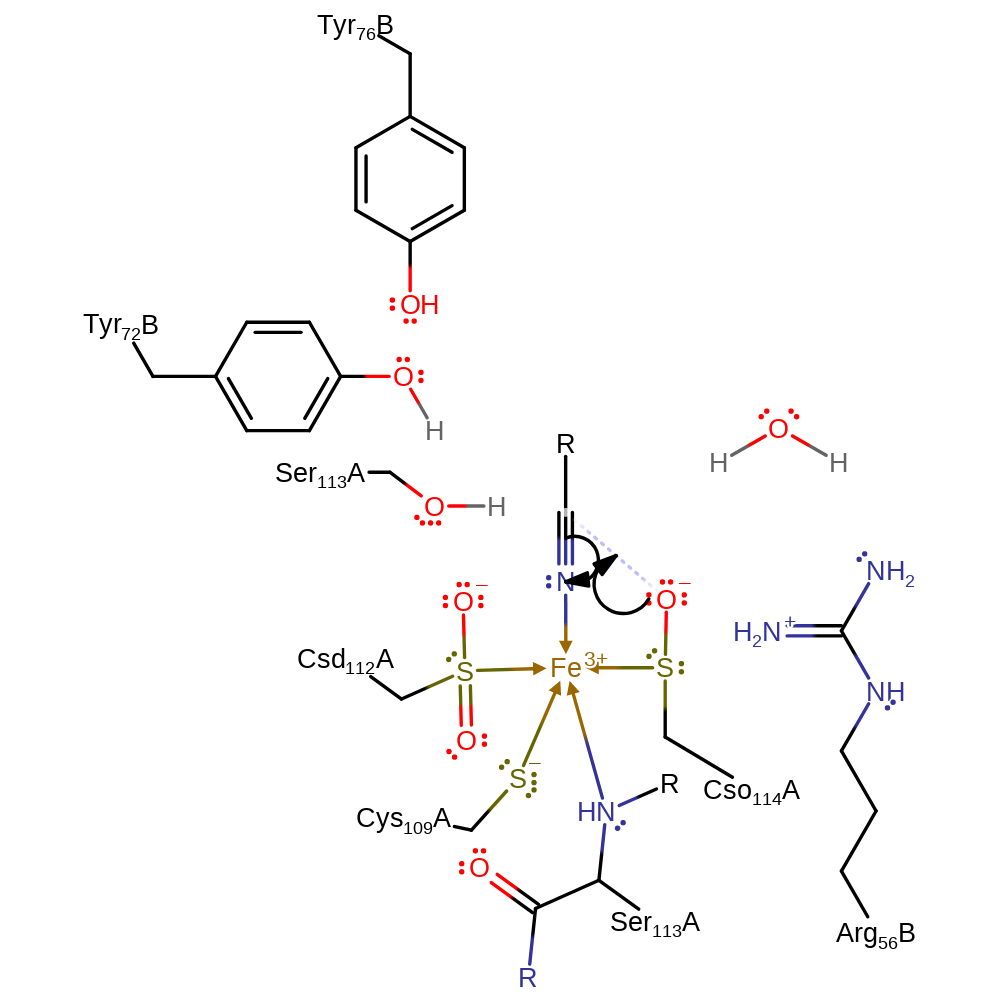
Step 1. The nitrile displaces a water molecule. The Cso hydroxide then initates a nucleophilic attack on the carbon of the nitrile.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg56B | electrostatic stabiliser |
| Cso114A (ptm) | nucleophile |
| Cys109A | metal ligand |
| Cso114A (ptm) | metal ligand |
| Csd112A (ptm) | metal ligand |
Chemical Components
ingold: bimolecular nucleophilic addition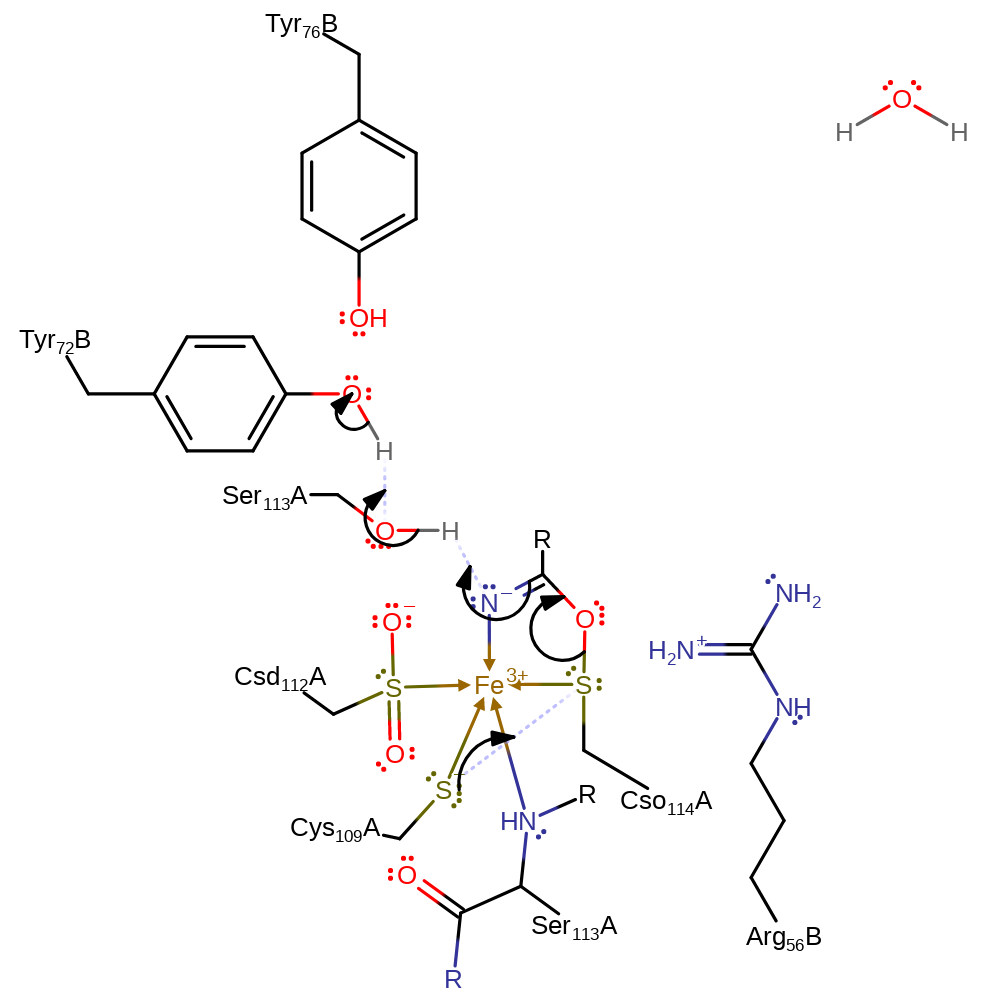
Step 2. Cys109A initiates a nucleophilic attack on the sulfur of Cso114, forming a disulfide bond and eliminating the nitrile intermediate. A proton is transferred from Tyr72B, via Ser113A, to the ngatvely charged nitrogen of the intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg56B | electrostatic stabiliser |
| Ser113A (main-N) | metal ligand |
| Cso114A (ptm) | nucleophile |
| Cys109A | metal ligand |
| Cso114A (ptm) | metal ligand |
| Csd112A (ptm) | metal ligand |
| Ser113A | proton relay |
| Tyr76B | increase acidity |
| Cso114A (ptm) | proton acceptor |
| Tyr72B | proton donor |
| Ser113A | proton acceptor, proton donor |
| Cys109A | nucleophile |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution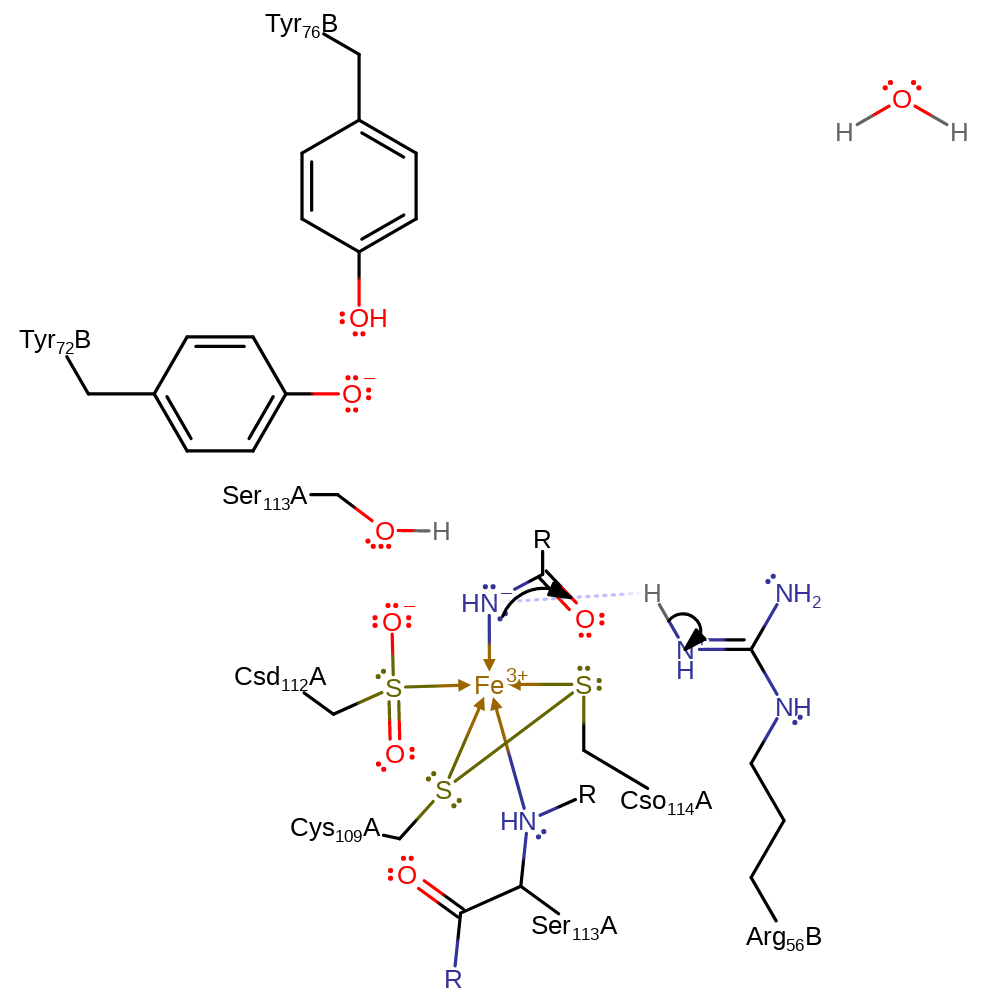
Step 3. The negatively charged nitrogen abstracts a proton from Arg56B.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser113A (main-N) | metal ligand |
| Cys109A | metal ligand |
| Csd112A (ptm) | metal ligand |
| Arg56B | proton donor |
Chemical Components
tautomerisation (not keto-enol), proton transfer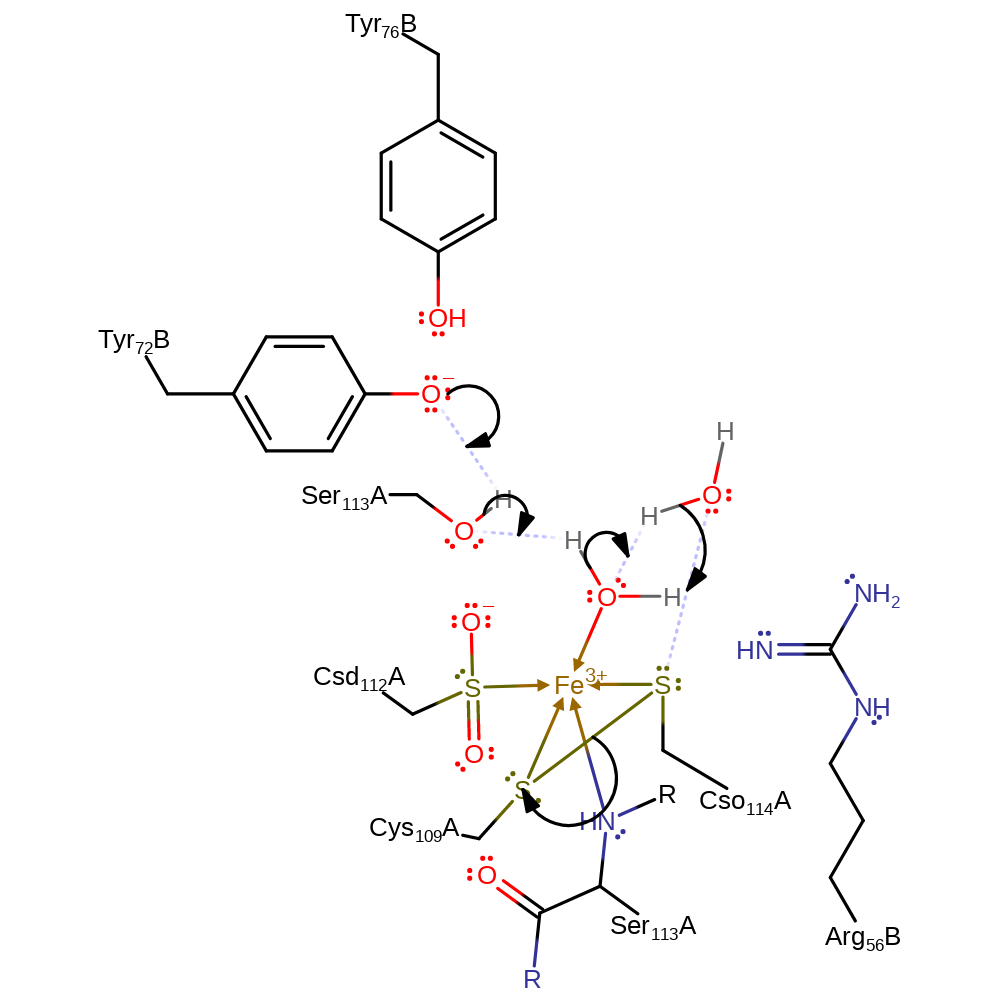
Step 4. Two water molecules enter the active site, one binds to the iron ion, displacing the substrate. Then, the negatively charged Tyrosine (Tyr72B) abstracts a proton Ser113A, which in turn abstracts a proton from the iron bound water. This then abstracts a proton from a second water molecule, which initiates a nucleophilic attack on the sulfur of Cso114, cleaving the disulfide bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg56B | electrostatic stabiliser |
| Ser113A (main-N) | metal ligand |
| Cys109A | metal ligand |
| Csd112A (ptm) | metal ligand |
| Cso114A (ptm) | metal ligand |
| Tyr76B | increase basicity |
| Ser113A | proton relay |
| Cys109A | nucleofuge |
| Tyr72B | proton acceptor |
| Ser113A | proton donor |
| Cso114A (ptm) | electrofuge |
| Ser113A | proton acceptor |
Chemical Components
ingold: bimolecular nucleophilic substitution, proton transfer
Step 5. It is unclear if this step occurs in a concerted manner with the previous one. The Arg56B abstracts a proton from the OH of Cso114A, regenerating the enzyme ground state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys109A | metal ligand |
| Cso114A (ptm) | metal ligand |
| Csd112A (ptm) | metal ligand |
| Cso114A (ptm) | proton donor |
| Arg56B | proton acceptor |
Chemical Components
proton transfer, native state of enzyme regeneratedIntroduction
The enzyme is activated through a photochemical activation of water in which a bound NO is replaced with HO-. In this proposal, a water molecule (activated by iron, and possibly the PTM CSO residue) initiates a nucleophilic attack on the nitrile substrate. The imidic acid intermediate is displaced by an incoming water molecule and undergoes tautomerisation without the help of the active site.
Catalytic Residues Roles
| UniProt | PDB* (2ahj) | ||
| Arg56 | Arg56B | Helps stabilise the reactive intermediates, and maintain the correct protonation state of the active site. | electrostatic stabiliser |
| Cys110, Cys115 (ptm), Cys113 (ptm), Ser114 (main-N) | Cys109A, Cso114A (ptm), Csd112A (ptm), Ser113A (main-N) | Forms part of the iron binding site. | metal ligand |
| Ser114, Tyr72 | Ser113A, Tyr72B | Forms part of the Ser-Tyr-Tyr catalytic triad, acts as a general acid/base. | proton relay, proton acceptor, proton donor |
| Tyr76 | Tyr76B | Forms part of the Ser-Tyr-Tyr catalytic triad, responsible for activating Tyr72 and Ser113 as general acid/bases. | increase basicity, increase acidity |
Chemical Components
bimolecular nucleophilic addition, proton transfer, assisted tautomerisation (not keto-enol)References
- Hashimoto K et al. (2008), J Biol Chem, 283, 36617-36623. Catalytic Mechanism of Nitrile Hydratase Proposed by Time-resolved X-ray Crystallography Using a Novel Substrate, tert-Butylisonitrile. DOI:10.1074/jbc.m806577200. PMID:18948265.
- Light KM et al. (2015), Chem Sci, 6, 6280-6294. Spectroscopic and computational studies of nitrile hydratase: insights into geometric and electronic structure and the mechanism of amide synthesis. DOI:10.1039/c5sc02012c. PMID:26508996.
- Martinez S et al. (2015), J Biol Inorg Chem, 20, 885-894. Analyzing the catalytic role of active site residues in the Fe-type nitrile hydratase from Comamonas testosteroni Ni1. DOI:10.1007/s00775-015-1273-3. PMID:26077812.
- MacDonald CA et al. (2015), Comput Theor Chem, 1070, 48-54. Competing nitrile hydratase catalytic mechanisms: Is cysteine-sulfenic acid acting as a nucleophile? DOI:10.1016/j.comptc.2015.07.010.
- Kayanuma M et al. (2015), Chem Phys Lett, 623, 8-13. A QM/MM study of the initial steps of catalytic mechanism of nitrile hydratase. DOI:10.1016/j.cplett.2015.01.039.
- Gumataotao N et al. (2013), J Biol Chem, 288, 15532-15536. Identification of an Active Site-bound Nitrile Hydratase Intermediate through Single Turnover Stopped-flow Spectroscopy. DOI:10.1074/jbc.m112.398909. PMID:23589282.
- Yamanaka Y et al. (2010), J Biol Inorg Chem, 15, 655-665. Kinetic and structural studies on roles of the serine ligand and a strictly conserved tyrosine residue in nitrile hydratase. DOI:10.1007/s00775-010-0632-3. PMID:20221653.
- Song L et al. (2007), Biochem Biophys Res Commun, 362, 319-324. High resolution X-ray molecular structure of the nitrile hydratase from Rhodococcus erythropolis AJ270 reveals posttranslational oxidation of two cysteines into sulfinic acids and a novel biocatalytic nitrile hydration mechanism. DOI:10.1016/j.bbrc.2007.07.184. PMID:17716629.
- Mitra S et al. (2007), J Biol Chem, 282, 7397-7404. Unraveling the Catalytic Mechanism of Nitrile Hydratases. DOI:10.1074/jbc.m604117200. PMID:17150969.
- Endo I et al. (2001), J Inorg Biochem, 83, 247-253. Fe-type nitrile hydratase. DOI:10.1016/s0162-0134(00)00171-9. PMID:11293544.
- Murakami T et al. (2000), Protein Sci, 9, 1024-1030. Post-translational modification is essential for catalytic activity of nitrile hydratase. DOI:10.1110/ps.9.5.1024. PMID:10850812.
- Piersma SR et al. (2000), J Inorg Biochem, 80, 283-288. Arginine 56 mutation in the β subunit of nitrile hydratase: importance of hydrogen bonding to the non-heme iron center. DOI:10.1016/s0162-0134(00)00076-3. PMID:11001100.
- Kobayashi M et al. (2000), Curr Opin Chem Biol, 4, 531-547. Nitrile Hydrolases. DOI:10.1007/1-4020-5377-0_30. PMID:10679370.
- Endo I et al. (1999), Trends Biotechnol, 17, 244-248. An enzyme controlled by light: the molecular mechanism of photoreactivity in nitrile hydratase. DOI:10.1016/s0167-7799(99)01303-7. PMID:10354562.
- Nagashima S et al. (1998), Nat Struct Biol, 5, 347-351. Novel non-heme iron center of nitrile hydratase with a claw setting of oxygen atoms. DOI:10.1038/nsb0598-347. PMID:9586994.
- Huang W et al. (1997), Structure, 5, 691-699. Crystal structure of nitrile hydratase reveals a novel iron centre in a novel fold. DOI:10.1016/s0969-2126(97)00223-2. PMID:9195885.
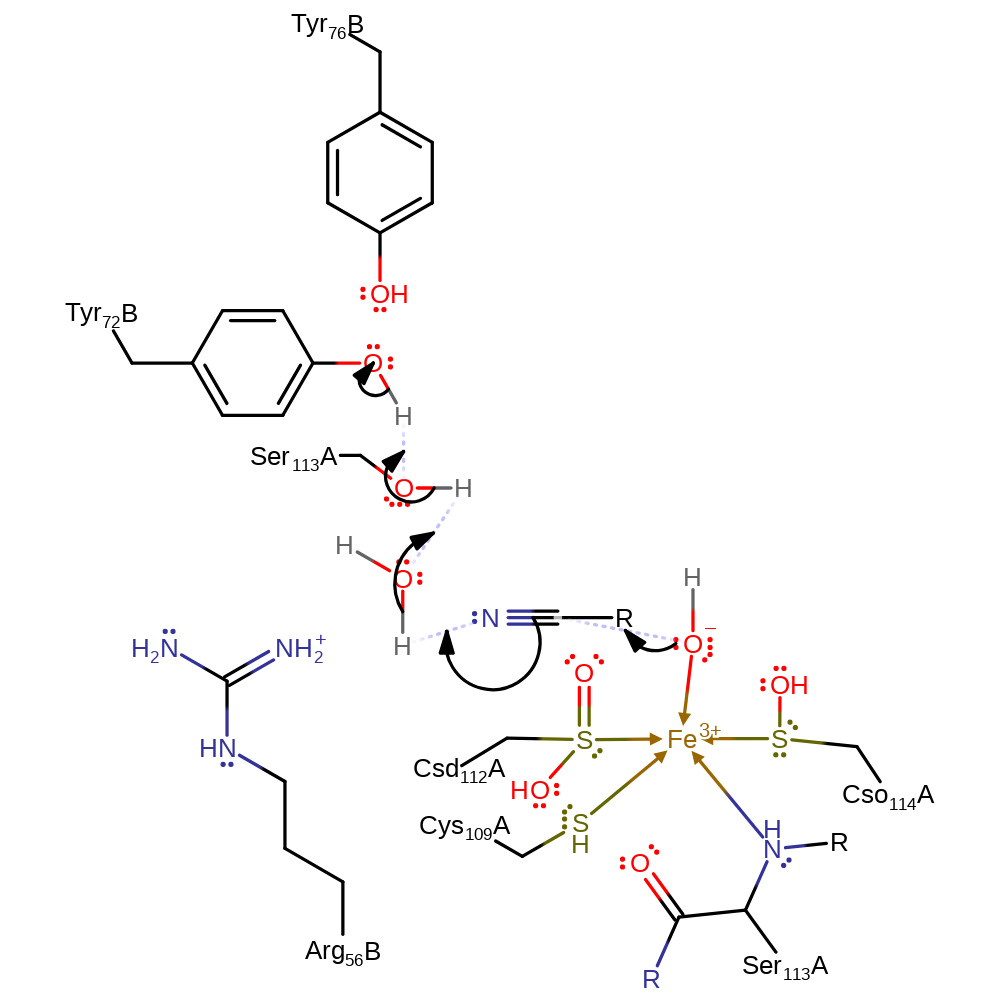
Step 1. Iron activated water adds to the carbon of the nitrile molecule in a nucleophilic attack. The nitrogen of the nitrile group abstracts a proton from the incoming water molecule through a proton relay with Ser113A and Tyr72B.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser113A (main-N) | metal ligand |
| Arg56B | electrostatic stabiliser |
| Cys109A | metal ligand |
| Csd112A (ptm) | metal ligand |
| Cso114A (ptm) | metal ligand |
| Tyr76B | increase acidity |
| Ser113A | proton relay, proton donor, proton acceptor |
| Tyr72B | proton donor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer
Step 2. The imidic acid intermediate undergoes tautomerisation to form the amide product. The Tyr72 abstracts a proton from the incoming water (which displaces the product) via Ser113 to regenerate the active site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser113A (main-N) | metal ligand |
| Cys109A | metal ligand |
| Csd112A (ptm) | metal ligand |
| Cso114A (ptm) | metal ligand |
| Tyr76B | increase basicity |
| Ser113A | proton relay |
| Tyr72B | proton acceptor |
| Ser113A | proton donor, proton acceptor |



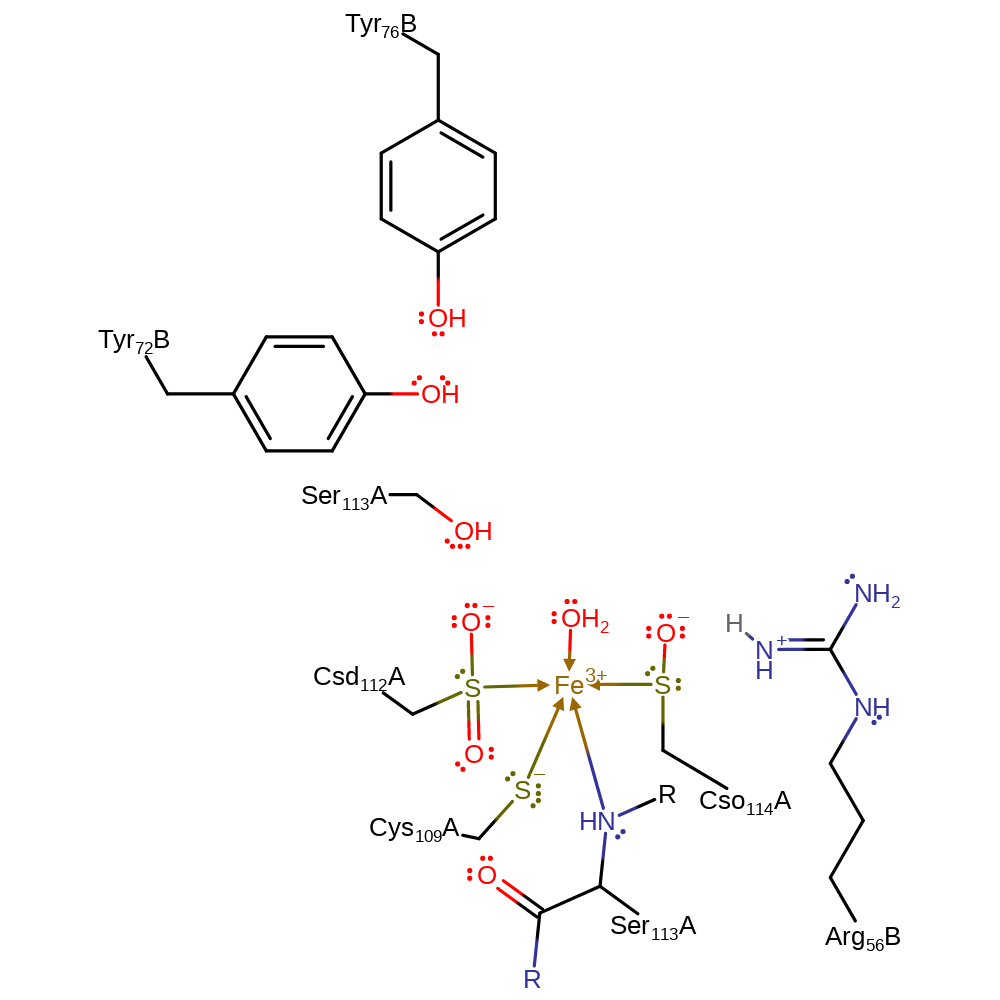 Download:
Download: 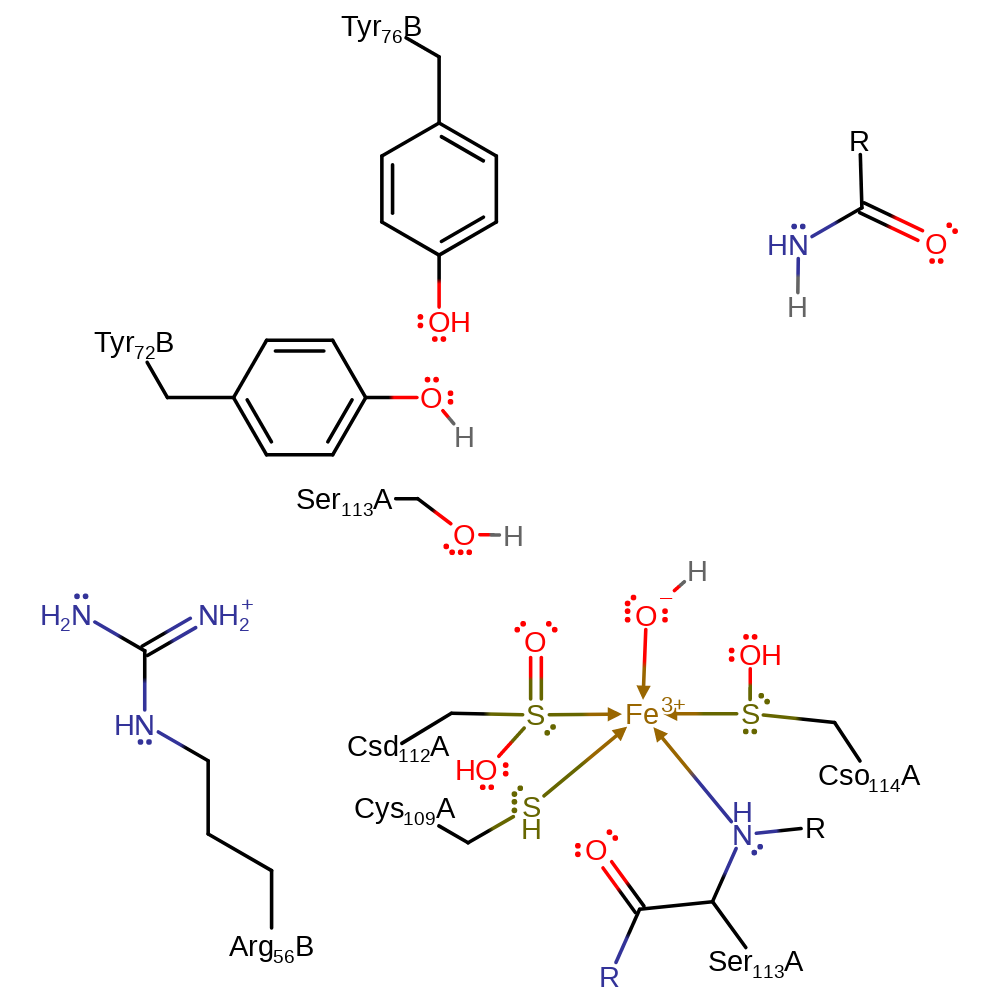 Download:
Download: