 |
PDBsum entry 3std

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.1.94
- scytalone dehydratase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
scytalone = 1,3,8-trihydroxynaphthalene + H2O
|
 |
 |
 |
 |
 |
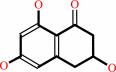 scytalone
scytalone
|
=
|
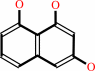 1,3,8-trihydroxynaphthalene
1,3,8-trihydroxynaphthalene
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
37:17735-17744
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure-based design of potent inhibitors of scytalone dehydratase: displacement of a water molecule from the active site.
|
|
J.M.Chen,
S.L.Xu,
Z.Wawrzak,
G.S.Basarab,
D.B.Jordan.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Scytalone dehydratase (SD) is a molecular target of inhibitor design efforts
aimed at protecting rice plants from the fungal disease caused by Magnaporthe
grisea. As determined from X-ray diffraction data of an SD-inhibitor complex
[Lundqvist et al. (1994) Structure (London) 2, 937-944], there is an extended
hydrogen-bonding network between protein side chains, the inhibitor, and two
bound water molecules. From models of SD complexed to quinazoline and
benztriazine inhibitors, a new class of potent SD inhibitors involving the
displacement of an active-site water molecule were designed. We were able to
increase inhibitory potency by synthesizing compounds with a nitrile
functionality displayed into the space occupied by one of the crystallographic
water molecules. Sixteen inhibitors are compared. The net conversion of potent
quinazoline and benztriazine inhibitors to cyanoquinolines and cyanocinnolines
increased binding potency 2-20-fold. Replacement of the nitrile with a hydrogen
atom lowered binding affinity 100-30,000-fold. X-ray crystallographic data at
1.65 A resolution on a SD-inhibitor complex confirmed that the nitrile
functionality displaced the water molecule as intended and that a favorable
orientation was created with tyrosines 30 and 50 which had been part of the
hydrogen-bonding network with the water molecule. Additional data on inhibitors
presented herein reveals the importance of two hydrogen-bonding networks toward
inhibitory potency: one between Asn131 and an appropriately positioned inhibitor
heteroatom and one between a bound water molecule and a second inhibitor
heteroatom.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Bissantz,
B.Kuhn,
and
M.Stahl
(2010).
A medicinal chemist's guide to molecular interactions.
|
| |
J Med Chem,
53,
5061-5084.
|
 |
|
|
|
|
 |
J.Michel,
J.Tirado-Rives,
and
W.L.Jorgensen
(2009).
Prediction of the water content in protein binding sites.
|
| |
J Phys Chem B,
113,
13337-13346.
|
 |
|
|
|
|
 |
J.Michel,
J.Tirado-Rives,
and
W.L.Jorgensen
(2009).
Energetics of displacing water molecules from protein binding sites: consequences for ligand optimization.
|
| |
J Am Chem Soc,
131,
15403-15411.
|
 |
|
|
|
|
 |
S.Wong,
R.E.Amaro,
and
J.A.McCammon
(2009).
MM-PBSA Captures Key Role of Intercalating Water Molecules at a Protein-Protein Interface.
|
| |
J Chem Theory Comput,
5,
422-429.
|
 |
|
|
|
|
 |
D.Katagiri,
H.Fuji,
S.Neya,
and
T.Hoshino
(2008).
Ab initio protein structure prediction with force field parameters derived from water-phase quantum chemical calculation.
|
| |
J Comput Chem,
29,
1930-1944.
|
 |
|
|
|
|
 |
S.K.Srivastava,
D.Dube,
V.Kukshal,
A.K.Jha,
K.Hajela,
and
R.Ramachandran
(2007).
NAD+-dependent DNA ligase (Rv3014c) from Mycobacterium tuberculosis: novel structure-function relationship and identification of a specific inhibitor.
|
| |
Proteins,
69,
97.
|
 |
|
|
|
|
 |
Z.Li,
and
T.Lazaridis
(2007).
Water at biomolecular binding interfaces.
|
| |
Phys Chem Chem Phys,
9,
573-581.
|
 |
|
|
|
|
 |
A.T.García-Sosa,
and
R.L.Mancera
(2006).
The effect of a tightly bound water molecule on scaffold diversity in the computer-aided de novo ligand design of CDK2 inhibitors.
|
| |
J Mol Model,
12,
422-431.
|
 |
|
|
|
|
 |
K.L.White,
J.M.Chen,
N.A.Margot,
T.Wrin,
C.J.Petropoulos,
L.K.Naeger,
S.Swaminathan,
and
M.D.Miller
(2004).
Molecular mechanisms of tenofovir resistance conferred by human immunodeficiency virus type 1 reverse transcriptase containing a diserine insertion after residue 69 and multiple thymidine analog-associated mutations.
|
| |
Antimicrob Agents Chemother,
48,
992.
|
 |
|
|
|
|
 |
L.Xu,
C.Li,
A.J.Olson,
and
I.A.Wilson
(2004).
Crystal structure of avian aminoimidazole-4-carboxamide ribonucleotide transformylase in complex with a novel non-folate inhibitor identified by virtual ligand screening.
|
| |
J Biol Chem,
279,
50555-50565.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Lim,
and
N.C.Strynadka
(2002).
Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus.
|
| |
Nat Struct Biol,
9,
870-876.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.A.Fritz,
D.Tondi,
J.S.Finer-Moore,
M.P.Costi,
and
R.M.Stroud
(2001).
Predicting and harnessing protein flexibility in the design of species-specific inhibitors of thymidylate synthase.
|
| |
Chem Biol,
8,
981-995.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.B.Jordan,
G.S.Basarab,
J.J.Steffens,
R.S.Schwartz,
and
J.G.Doughty
(2000).
Tight binding inhibitors of scytalone dehydratase: effects of site-directed mutations.
|
| |
Biochemistry,
39,
8593-8602.
|
 |
|
|
|
|
 |
D.B.Jordan,
and
G.S.Basarab
(2000).
Binding dynamics of two water molecules constrained within the scytalone dehydratase binding pocket.
|
| |
Bioorg Med Chem Lett,
10,
23-26.
|
 |
|
|
|
|
 |
D.B.Jordan,
Y.J.Zheng,
B.A.Lockett,
and
G.S.Basarab
(2000).
Stereochemistry of the enolization of scytalone by scytalone dehydratase.
|
| |
Biochemistry,
39,
2276-2282.
|
 |
|
|
|
|
 |
D.I.Liao,
G.S.Basarab,
A.A.Gatenby,
and
D.B.Jordan
(2000).
Selection of a potent inhibitor of trihydroxynaphthalene reductase by sorting disease control data.
|
| |
Bioorg Med Chem Lett,
10,
491-494.
|
 |
|
|
|
|
 |
A.E.Nixon,
S.M.Firestine,
F.G.Salinas,
and
S.J.Benkovic
(1999).
Rational design of a scytalone dehydratase-like enzyme using a structurally homologous protein scaffold.
|
| |
Proc Natl Acad Sci U S A,
96,
3568-3571.
|
 |
|
|
|
|
 |
G.S.Basarab,
D.B.Jordan,
T.C.Gehret,
R.S.Schwartz,
and
Z.Wawrzak
(1999).
Design of scytalone dehydratase inhibitors as rice blast fungicides: derivatives of norephedrine.
|
| |
Bioorg Med Chem Lett,
9,
1613-1618.
|
 |
|
|
|
|
 |
L.D.Jennings,
Z.Wawrzak,
D.Amorose,
R.S.Schwartz,
and
D.B.Jordan
(1999).
A new potent inhibitor of fungal melanin biosynthesis identified through combinatorial chemistry.
|
| |
Bioorg Med Chem Lett,
9,
2509-2514.
|
 |
|
|
|
|
 |
Z.Wawrzak,
T.Sandalova,
J.J.Steffens,
G.S.Basarab,
T.Lundqvist,
Y.Lindqvist,
and
D.B.Jordan
(1999).
High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
|
| |
Proteins,
35,
425-439.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

