 |
PDBsum entry 1qcc

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.7
- adenine phosphoribosyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Ribose activation
|
 |
 |
 |
 |
 |
Reaction:
|
 |
AMP + diphosphate = 5-phospho-alpha-D-ribose 1-diphosphate + adenine
|
 |
 |
 |
 |
 |
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
=
|
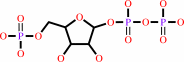 5-phospho-alpha-D-ribose 1-diphosphate
5-phospho-alpha-D-ribose 1-diphosphate
|
+
|
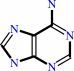 adenine
adenine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
EMBO J
18:3533-3545
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of adenine phosphoribosyltransferase from Leishmania donovani.
|
|
C.L.Phillips,
B.Ullman,
R.G.Brennan,
C.P.Hill.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The enzyme adenine phosphoribosyltransferase (APRT) functions to salvage adenine
by converting it to adenosine-5-monophosphate (AMP). APRT deficiency in humans
is a well characterized inborn error of metabolism, and APRT may contribute to
the indispensable nutritional role of purine salvage in protozoan parasites, all
of which lack de novo purine biosynthesis. We determined crystal structures for
APRT from Leishmania donovani in complex with the substrate adenine, the product
AMP, and sulfate and citrate ions that appear to mimic the binding of phosphate
moieties. Overall, these structures are very similar to each other, although the
adenine and AMP complexes show different patterns of hydrogen-bonding to the
base, and the active site pocket opens slightly to accommodate the larger AMP
ligand. Whereas AMP adopts a single conformation, adenine binds in two mutually
exclusive orientations: one orientation providing adenine-specific hydrogen
bonds and the other apparently positioning adenine for the enzymatic reaction.
The core of APRT is similar to that of other phosphoribosyltransferases,
although the adenine-binding domain is quite different. A C-terminal extension,
unique to Leishmania APRTs, extends an extensive dimer interface by wrapping
around the partner molecule. The active site involves residues from both
subunits of the dimer, indicating that dimerization is essential for catalysis.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 7.
Figure 7 Schematic representation of contacts to AMP. Hydrogen
bonds and metal–ligand contacts are indicated with a dotted
line and defined as for Figure 8.
|
 |
Figure 10.
Figure 10 Superposition of APRT active site structures.
AMP–APRT red; apo-AS–APRT green; Ade–APRT A orientation
light blue; Ade–APRT B orientation dark blue.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Macmillan Publishers Ltd:
EMBO J
(1999,
18,
3533-3545)
copyright 1999.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Kukimoto-Niino,
R.Shibata,
K.Murayama,
H.Hamana,
M.Nishimoto,
Y.Bessho,
T.Terada,
M.Shirouzu,
S.Kuramitsu,
and
S.Yokoyama
(2005).
Crystal structure of a predicted phosphoribosyltransferase (TT1426) from Thermus thermophilus HB8 at 2.01 A resolution.
|
| |
Protein Sci,
14,
823-827.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.H.Rehse,
and
T.H.Tahirov
(2005).
Crystal structure of a purine/pyrimidine phosphoribosyltransferase-related protein from Thermus thermophilus HB8.
|
| |
Proteins,
61,
658-665.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.H.el Kouni
(2003).
Potential chemotherapeutic targets in the purine metabolism of parasites.
|
| |
Pharmacol Ther,
99,
283-309.
|
 |
|
|
|
|
 |
S.C.Sinha,
J.Krahn,
B.S.Shin,
D.R.Tomchick,
H.Zalkin,
and
J.L.Smith
(2003).
The purine repressor of Bacillus subtilis: a novel combination of domains adapted for transcription regulation.
|
| |
J Bacteriol,
185,
4087-4098.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.E.Sarver,
and
C.C.Wang
(2002).
The adenine phosphoribosyltransferase from Giardia lamblia has a unique reaction mechanism and unusual substrate binding properties.
|
| |
J Biol Chem,
277,
39973-39980.
|
 |
|
|
|
|
 |
A.Kadziola,
J.Neuhard,
and
S.Larsen
(2002).
Structure of product-bound Bacillus caldolyticus uracil phosphoribosyltransferase confirms ordered sequential substrate binding.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
936-945.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Bashor,
J.M.Denu,
R.G.Brennan,
and
B.Ullman
(2002).
Kinetic mechanism of adenine phosphoribosyltransferase from Leishmania donovani.
|
| |
Biochemistry,
41,
4020-4031.
|
 |
|
|
|
|
 |
M.A.Schumacher,
C.J.Bashor,
M.H.Song,
K.Otsu,
S.Zhu,
R.J.Parry,
B.Ullman,
and
R.G.Brennan
(2002).
The structural mechanism of GTP stabilized oligomerization and catalytic activation of the Toxoplasma gondii uracil phosphoribosyltransferase.
|
| |
Proc Natl Acad Sci U S A,
99,
78-83.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Allen,
W.Qin,
F.Moreau,
and
B.Moffatt
(2002).
Adenine phosphoribosyltransferase isoforms of Arabidopsis and their potential contributions to adenine and cytokinin metabolism.
|
| |
Physiol Plant,
115,
56-68.
|
 |
|
|
|
|
 |
O.Mayans,
A.Ivens,
L.J.Nissen,
K.Kirschner,
and
M.Wilmanns
(2002).
Structural analysis of two enzymes catalysing reverse metabolic reactions implies common ancestry.
|
| |
EMBO J,
21,
3245-3254.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.Shi,
A.E.Sarver,
C.C.Wang,
K.S.Tanaka,
S.C.Almo,
and
V.L.Schramm
(2002).
Closed site complexes of adenine phosphoribosyltransferase from Giardia lamblia reveal a mechanism of ribosyl migration.
|
| |
J Biol Chem,
277,
39981-39988.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.R.Bonner,
J.N.D'Elia,
B.K.Billips,
and
R.L.Switzer
(2001).
Molecular recognition of pyr mRNA by the Bacillus subtilis attenuation regulatory protein PyrR.
|
| |
Nucleic Acids Res,
29,
4851-4865.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

