
If each human cell is a galaxy, the creation of a new vesicle is like the birth of a new planet. The ethereal beauty of this month’s artwork gives endocytosis a mystical quality. Endocytosis is fundamental to us in the same way as eating and drinking are fundamental to our continued survival. Critical nutrients, hormones and receptors can only get where they are needed via endocytosis. Endocytosis is the process where the cell membrane folds in on itself and pinches together to release a membrane-encased package, almost as though the cell is ‘blowing bubbles’ internally. With endocytosis, things that were previously outside the cell become encased in membrane-packages that are now inside the cell.
This month’s artwork highlights in different colours the different subunits of the adaptin protein complex. This complex associates with the membrane and with membrane-embedded proteins. The association of the adaptin complex subunits with each other and the membrane are key coordinating steps for a certain type of endocytosis. In this article we will explore the adaptin protein complex, as well as another protein called clathrin. These proteins work together to reshape small sections of cell membrane into 'bubbles' to carry nutrients and hormones inside the cell.
Different types of endocytosis
When we think about endocytosis, the most common type we usually picture first is when a large particle, such as a bacterial cell, is engulfed by a human macrophage, resulting in the bacterial cell being encased in a vesicle named “phagosome”. This vesicle usually has a size ranging from 0.5 to 5-microns. There is a more specialised name for this endocytosis process, phagocytosis, and the vesicle is subsequently acidified to aid in the degradation of the bacterial cell.
Another type of endocytosis is clathrin-mediated endocytosis which was first observed over 50 years ago using electron microscopy. For this process, the clathrin protein chains form a cage-like lattice or scaffold that reshapes the membrane into a vesicle (PDB ID 3IYV, Figure 1). Clathrin-coated vesicles have sizes typically ranging from 40 to 100 nm (0.04 to 0.10 microns), thus these vesciles are 5 to 100 times smaller than phagosomes. Clathrin does not directly interact with membranes. Clathrin interacts with membranes via an adaptor protein complex named "adaptin protein complex".

Figure 1.
Clathrin barrel with dimenisions similar to clathrin-coated vesicles present in our cells.
The hexagonal clathrin barrel (PDB ID 3IYV) is over 70 nm in diameter and over 80 nm in length (0.07 by 0.08 microns), with D6 symmetry. The barrel involves 216 clathrin protein chains. Clathrin has two types of protein chains referred to as heavy chains (green) and light chains (orange). The clathrin protein chains in this structure are from cow (Bos taurus). Image was generated using Mol* (molstar.org/viewer).
Clathrin-coated vesicles carry components within the trans-Golgi network, between the trans-Golgi network and endosomes, but also from outside the cell to cellular organelles, including Golgi bodies and endosomes. The cage-like scaffold of clathrin is composed of triskelion units (PDB ID 3LVH, Figure 2A), that are reminiscent of the Isle of Man flag (Figure 2B). The clathrin triskelion units are like Lego blocks and can come together in slightly different ways to drive the formation of different membrane shapes and different-sized vesicles.

Figure 2.
The clathrin units that come together to form the clathrin barrel and other clathrin scaffold shapes.
(A) The clathrin triskelion unit is shown with spaced-filled spheres representing the atoms (PDB ID 3LVH). The clathrin protein chains in this structure are from cow (Bos taurus). (B) The flag for the Isle of Man also has a triskelion which is composed of three armoured legs with golden spurs. (C) Clathrin triskelion units shown with cartoon rather than space-filled representation. The clathrin triskelion is composed of three clathrin protein heavy chains (green; length of 20 nm) and three clathrin protein light chains (orange; length of 12 nm). Image was generated using Mol* (molstar.org/viewer).
The multi-subunit, multi-domain adaptin complex
Adaptin complexes are hetero-oligmers, i.e. they are always composed of multiple protein chains with different amino acid sequences. For adaptin complexes there are usually two larger subunits named alpha (α) and beta (β), and two smaller subunits called mu (μ; ∼50 kDa) and sigma (σ; 17–20 kDa). The structure in the artwork (PDB ID 2VGL) represents a breakthrough in our understanding of how these different protein subunits interact with each other.
Prior to the structure in the artwork (PDB ID 2VGL), there had been studies using electron microscopes, where adaptins appeared on the end of the clathrin triskelton. In these previous studies adaptins appeared as a large central brick-like core joined by flexible linkers to small “appendage” or “ear” domains that were attached to clathrin. These studies gave no details of the interactions between the adaptin subunits.
Larger adaptin subunits are composed of a "trunk" domain and the flexible linker and small “appendage” or “ear” domains. In order to get structural insight at higher resolution, the flexible linkers and the small “appendage” domains were excluded and only the "trunk" domain of the larger adaptin subunits (the α and β subunits) were included in thee strucrture and it was called the "adaptin complex core".
The structure featured in the artwork (PDB ID 2VGL) is the AP2 adaptin protein complex core and contains a bound chemical (inositol hexakisphosphate; CC ID IHP). The bound chemical is shown in Figure 3 in red, with space-filled spheres representing each atom. This compound can be considered analogous to the headgroups in inositol phospholipids (phosphatidylinositol; PtdIns) which are found in the cell membrane. Phospholipids position their headgroups on the membrane surface. The binding of this compound at this site in the adpatin protein complex helps indicate which side of the protein complex is orientated towards the cell membrane.
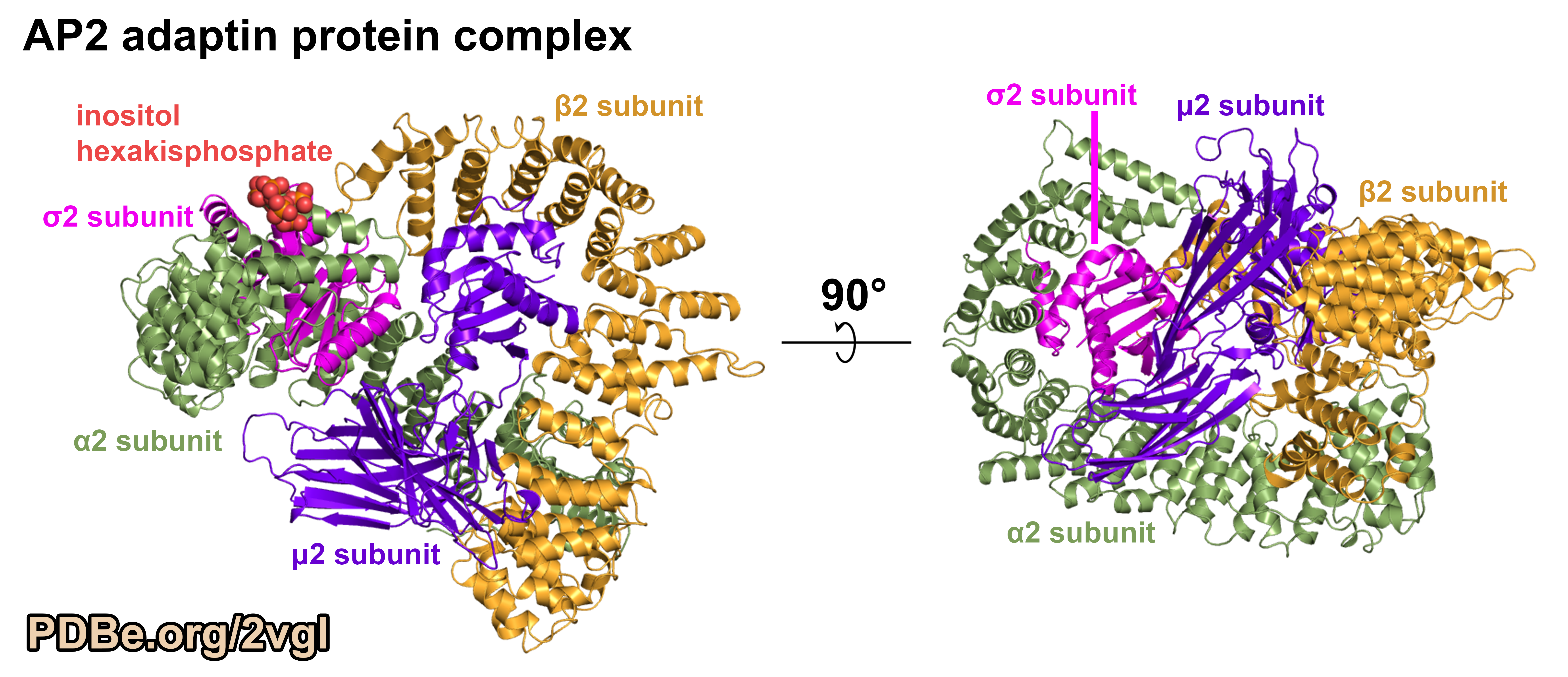
Figure 3.
AP2 adaptin complex core structure (PDB ID 2VGL) shows the details of the interactions between the different protein chains.
The four protein chains are the alpha-2 (α2; green), beta-2 (β2; orange), sigma-2 (σ2; pink), mu-2 (μ2; purple) subunits. This structure includes the complete μ2 and σ2 subunits, but only the "trunk" domains of the α2 and β2 subunits. It was solved by X-ray diffraction to a resolution of 2.6 Angstroms. The α2 and μ2 subunits have the protein sequence from rats (Rattus norvegicus), the σ2 subunit is from mice (Mus musculus), and the β2 subunit is from humans (Homo sapiens). Preparing proteins with sequences from different organisms is often part of the process when determining new structures because not all proteins from all organisms are found to behave well in the laboratory. Image was generated using PyMOL (pymol.org).
The surprising similarity between adaptin subunits
The AP2 adaptin complex core structure revealed the smallest subunit, σ2 subunit, and the N-terminal region of the second smallest subunit, μ2 subunit, have the same structural-fold. If one compares the two halves of the complex, that is the σ2:α2 heterodimer and the μ2:β2 heterodimer, by superimposing σ2 subunit on the N-terminal region of the μ2 subunit, more similarity between the subunits becomes apparent. The alpha-helices of the larger subunits, α2 subunit and β2 subunit, occur in similar positions relative to their interactions with the smaller adaptin subunits (PDB ID 2VGL, Figure 4). This pseudo-symmetry is surprising given the protein complex is composed of four different amino acid sequences.
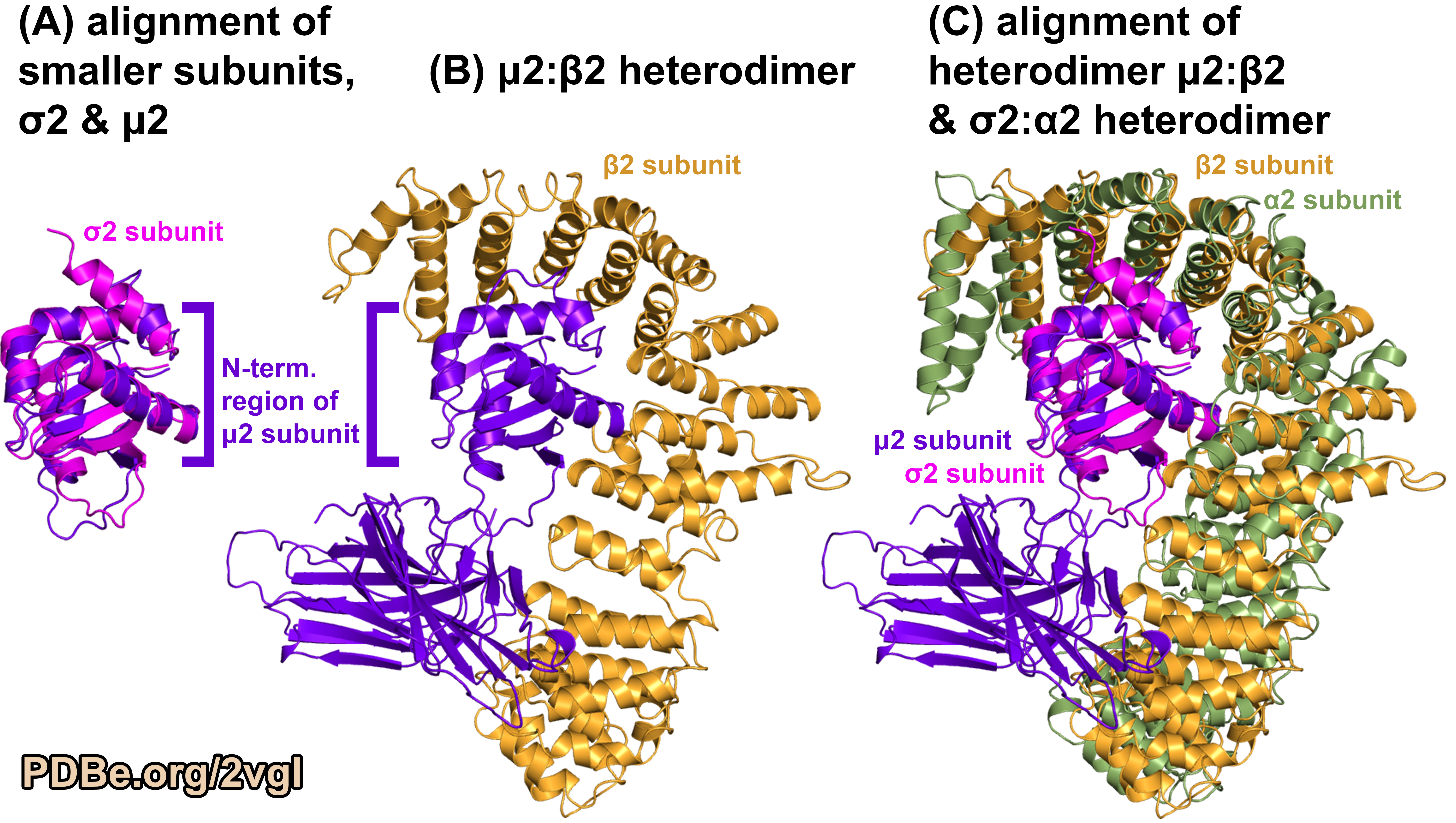
Figure 4.
Comparing the different subunits in the AP2 adaptin complex (PDB ID 2VGL).
(A) Structural superposition of the shared structured fold component from the smaller subunits of the protein complex. (B) One half of the protein complex, the μ2:β2 heterodimer (a larger and smaller subunit). (C) Structural superposition of the two halves of the protein complex using the shared structural-fold component from the smaller subunits. Image was generated using PyMOL (pymol.org).
More insight from adaptin structure
The C-terminal region of μ2 subunit is composed of only beta-sheets which is distinctive when contrasted with the structural fold of the other components in the adaptin complex core (Figure 4). Subsequent structures have revealed that the first structure of the AP2 adaptin complex core (PDB ID 2VGL) is a ‘closed’ conformation, and the binding of certain peptides and other proteins results in a more 'open’ conformation (e.g. PDB ID 2XA7). The C-terminal region of μ2 subunit has been observed to adopt a different position relative to the rest of the adaptin protein complex core (Figure 5).
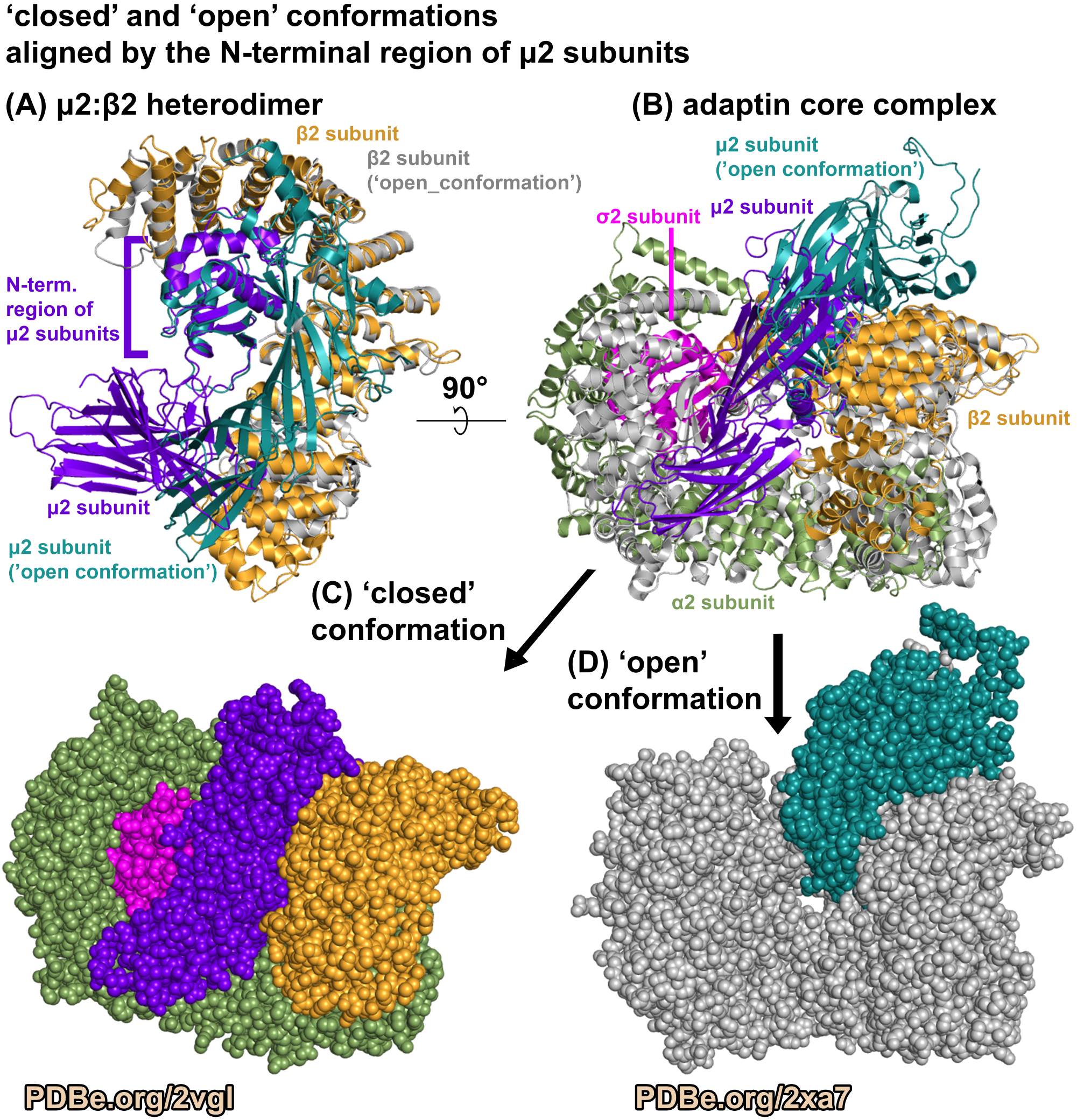
Figure 5.
Different conformations of the adaptin complex.
Comparing the AP2 adaptin core in a ‘closed’ conformation, (PDB ID 2VGL) and in an ‘open’ conformation, (PDB ID 2XA7). The ‘open’ conformation structure (PDB ID 2XA7) was solved by X-ray diffraction to a resolution of 3.1 Angstroms. The binding of a peptide causes the protein complex to have an 'open' conformation. Image was generated using PyMOL (pymol.org).
In the wwPDB database, thanks to the work from many scientists from different research groups, there are multiple structures of adaptin complex core, including AP1 and AP3 adaptin complexes.
Among the proteins identified that interact with adaptin are viral proteins (e.g. PDB IDs 4NEE, 6OWT, and 6URI) that interfere with the natural process to redirect ‘traffic’ to benefit virus production. Many of the structures in the wwPDB database that contain the adaptin complex core also contain different peptides, i.e. short protein stretches from other human and viral proteins. Many of the structures in the have the complex core in ‘closed’ or ‘open’ conformations. These structures help to identify the roles other proteins have in recruiting the adaptin complex core to the membrane and/or change its conformation and thus expand our understanding of how endocytosis is being coordinated at the molecular level. These interactions can either be triggering or blocking the subsequent steps that result in the formation of the clathrin-coated vesicles. Therefore, these other proteins, like the viral proteins, can either drive or block vesicle formation at the cell membrane or the membrane surface of cell organelles (e.g. endosome or Golgi body).
Expanding our understanding of these processes has the potential to illuminate new possibilities for antiviral drugs.
Genevieve Evans
About this Artwork
This artwork was created by Elspeth Owen from Perse High School. Elspeth chose to capture the beauty of this protein complex due to its interesting science and her personal connection to it. Elspeth shared: ‘Both my parents worked on the structure and I wanted to represent all their hard work in a way I enjoy.’ Elspeth experimented with contrasting colours from both the warm and cool colour families to see which would still produce pleasing colours if they blended during the printing process. There are two closeups of the protein structure from different angles to capture its intrinsic beauty. Elspeth shared: ‘This protein complex plays a key role in transporting the nutrients the cell needs through its surrounding membrane.’ Both in the background and at the bottom of the artwork aspects of the nutrients journey through the membrane are shown.
View the artwork in the virtual 2022 PDB Art exhibition.
Structures found in the images in this article
- Clathrin barrel (PDB ID 3IYV)
- Clathrin triskelion (PDB ID 3LVH)
- Adaptin complex core ‘closed’ conformation (PDB ID 2VGL)
- Adaptin complex core ‘open’ conformation (PDB ID 2XA7)
Sources
- PDB ID 3IYV: Molecular model for a complete clathrin lattice from electron cryo-microscopy
- PDB ID 3LVH: Conformation switching of clathrin light chain regulates clathrin lattice assembly
- PDB ID 2VGL: Molecular architecture and functional model of the endocytic AP2 complex
- PDB ID 2XA7: A Large-Scale Conformational Change Couples Membrane Recruitment to Cargo Binding in the AP2 Clathrin Adaptor Complex
More structures to explore
- Structures of adaptin complex core (including the larger subunit "trunk" domain),
as well as structures of "appendage" domain are listed on PDBe-KB pages for adaptin larger subunits: - Alphafold structure predictions for adaptin larger subunits, including "trunk" and "appendage" domains:
- Structure of adaptin larger subunit "appendage" domain interacting with clathrin protein chains:


