 |
PDBsum entry 4e7x
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Structural basis for the activity of a cytoplasmic RNA terminal u- transferase
|
|
Structure:
|
 |
Poly(a) RNA polymerase protein cid1. Chain: a, b, c, d. Synonym: caffeine-induced death protein 1. Engineered: yes
|
|
Source:
|
 |
Schizosaccharomyces pombe 972h-. Fission yeast. Organism_taxid: 284812. Strain: 972 / atcc 24843. Gene: cid1, spac19d5.03. Expressed in: escherichia coli. Expression_system_taxid: 511693.
|
|
Resolution:
|
 |
|
3.20Å
|
R-factor:
|
0.181
|
R-free:
|
0.213
|
|
|
Authors:
|
 |
L.A.Yates,S.Fleurdepine,O.S.Rissland,L.Decolibus,K.Harlos, C.J.Norbury,R.J.C.Gilbert
|
|
Key ref:
|
 |
L.A.Yates
et al.
(2012).
Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase.
Nat Struct Biol,
19,
782-787.
PubMed id:
|
 |
|
Date:
|
 |
|
19-Mar-12
|
Release date:
|
04-Jul-12
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O13833
(CID1_SCHPO) -
Terminal uridylyltransferase cid1 from Schizosaccharomyces pombe (strain 972 / ATCC 24843)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
405 a.a.
319 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.7.7.19
- polynucleotide adenylyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
RNA(n) + ATP = RNA(n)-3'-adenine ribonucleotide + diphosphate
|
 |
 |
 |
 |
 |
 RNA(n)
RNA(n)
|
+
|
 ATP
ATP
|
=
|
RNA(n)-3'-adenine ribonucleotide
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.7.7.52
- Rna uridylyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
RNA(n) + UTP = RNA(n)-3'-uridine ribonucleotide + diphosphate
|
 |
 |
 |
 |
 |
 RNA(n)
RNA(n)
|
+
|
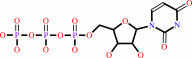 UTP
UTP
|
=
|
RNA(n)-3'-uridine ribonucleotide
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Nat Struct Biol
19:782-787
(2012)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase.
|
|
L.A.Yates,
S.Fleurdépine,
O.S.Rissland,
L.De Colibus,
K.Harlos,
C.J.Norbury,
R.J.Gilbert.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

