Adenine phosphoribosyltransferase
Adenine phosphoribosyltransferase (APRTase) catalyses the reversible Mg2+ dependent reaction of adenine with 5-phospho-alpha-D-ribosyl-1-pyrophosphate (PRPP) to produce AMP and pyrophosphate. This reaction is important in adenine salvage and recycling, and the enzyme is present in species ranging from bacteria to mammals. In humans, APRTase has the sole metabolic function of recycling adenine formed in the polyamine pathway, and the effects of APRTase deficiency are relatively mild. Some protozoan parasite, including Giardia lamblia, are deficient in de novo purine synthesis and so purine uptake from the host and the APRTase reaction are especially important in these organisms.
Reference Protein and Structure
- Sequence
-
Q967M2

 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Giardia intestinalis (Flagellated parasite)

- PDB
-
1l1r
- Crystal Structure of APRTase from Giardia lamblia Complexed with 9-deazaadenine, Mg2+ and PRPP
(1.95 Å)



- Catalytic CATH Domains
-
3.40.50.2020
 (see all for 1l1r)
(see all for 1l1r)
- Cofactors
- Magnesium(2+) (1)
Enzyme Reaction (EC:2.4.2.7)
Enzyme Mechanism
Introduction
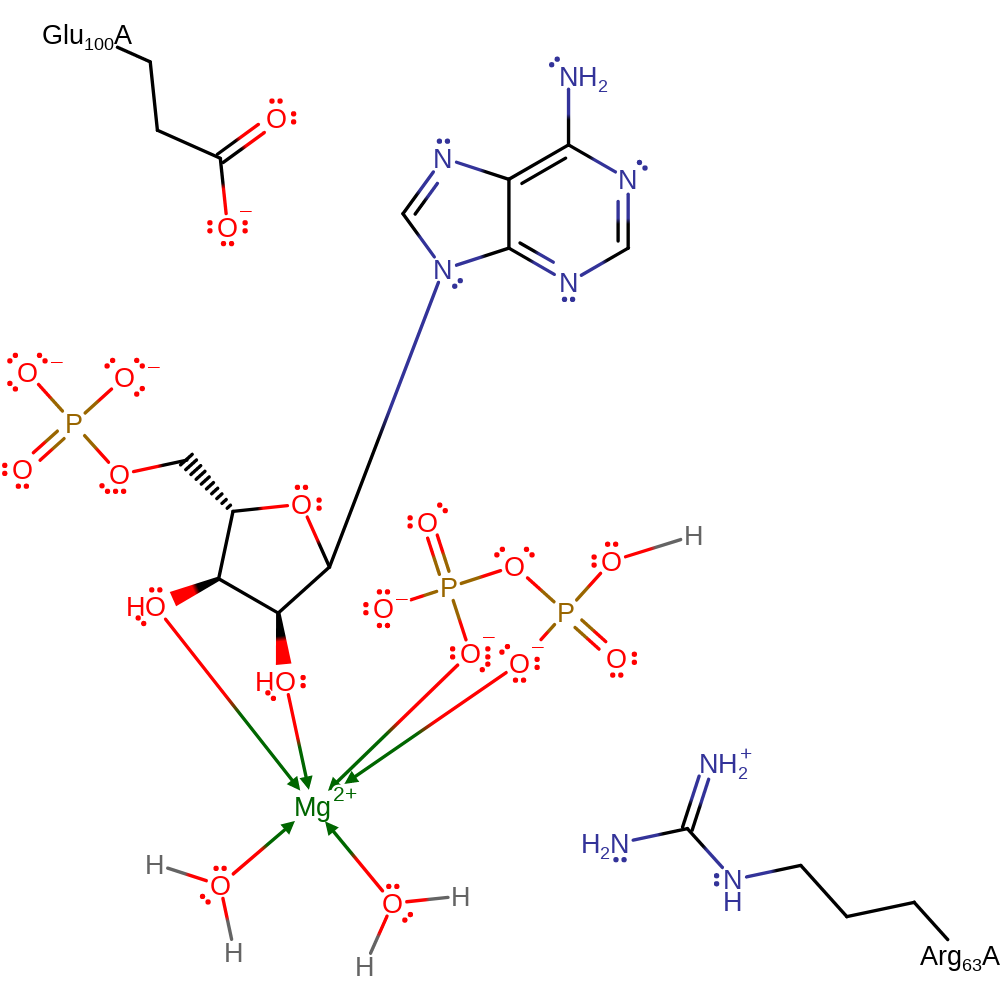
In the proposed Sn1 mechanism, Glu100 functions as a general base to deprotonate N7 of adenine. This enables the N7 of adenine to nucleophilically attack the electrophilic carbon. The reaction passes through an oxacarbenium-like transition state in which the Mg2+ ion and Arg 63 stabilise accumulation of negative charge on the pyrophosphate leaving group.
Catalytic Residues Roles
| UniProt | PDB* (1l1r) | ||
| Arg63 | Arg63A | Stabilises accumulation of negative charge on the pyrophosphate leaving group. | electrostatic stabiliser |
| Glu100 | Glu100A | Deprotonates N7 of adenine. | proton acceptor, proton donor |
Chemical Components
proton transfer, bimolecular nucleophilic substitution, overall reactant used, overall product formed, inferred reaction step, native state of enzyme regeneratedReferences
- Shi W et al. (2002), J Biol Chem, 277, 39981-39988. Closed Site Complexes of Adenine Phosphoribosyltransferase fromGiardia lamblia Reveal a Mechanism of Ribosyl Migration. DOI:10.1074/jbc.m205596200. PMID:12171925.
- Arco JD et al. (2017), Curr Pharm Des,Purine and Pyrimidine Phosphoribosytransferases: A versatile tool for enzymatic synthesis of nucleoside-5'-monophosphates. DOI:10.2174/1381612823666171017165707. PMID:29046144.
- Takahashi R et al. (2010), J Biochem, 147, 95-107. Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase. DOI:10.1093/jb/mvp152. PMID:19819904.
- Shi W et al. (2001), Biochemistry, 40, 10800-10809. Structural Analysis of Adenine Phosphoribosyltransferase fromSaccharomyces cerevisiae†,‡. DOI:10.1021/bi010465h. PMID:11535055.
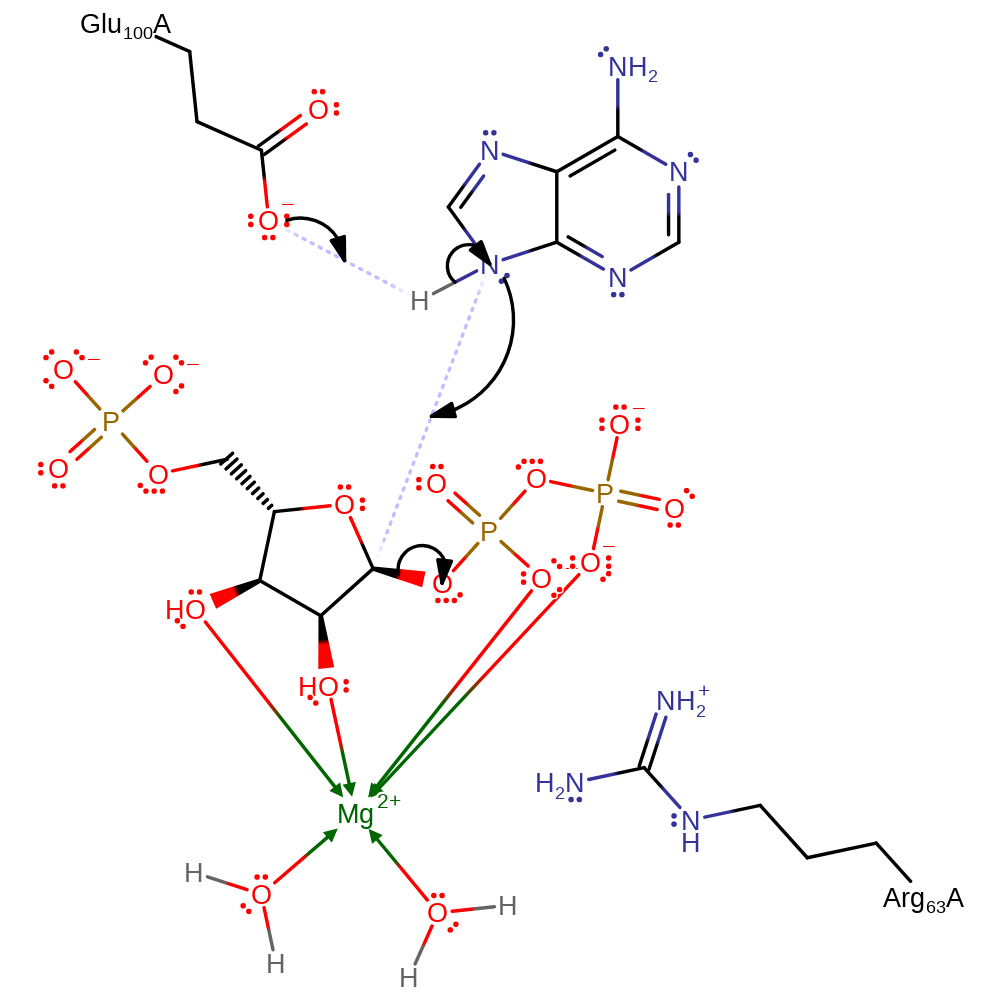
Step 1. Glu100 deprotonates N7 of adenine, increasing the base's nucleophilicity to attack the anomeric carbon on the ribose sugar in an SN1 like reaction with an oxocarbeniom ion like transition state which is stabilised by Arg63.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg63A | electrostatic stabiliser |
| Glu100A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, overall reactant usedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu100A | proton donor |
Chemical Components
overall product formed, proton transfer, inferred reaction step, native state of enzyme regeneratedIntroduction
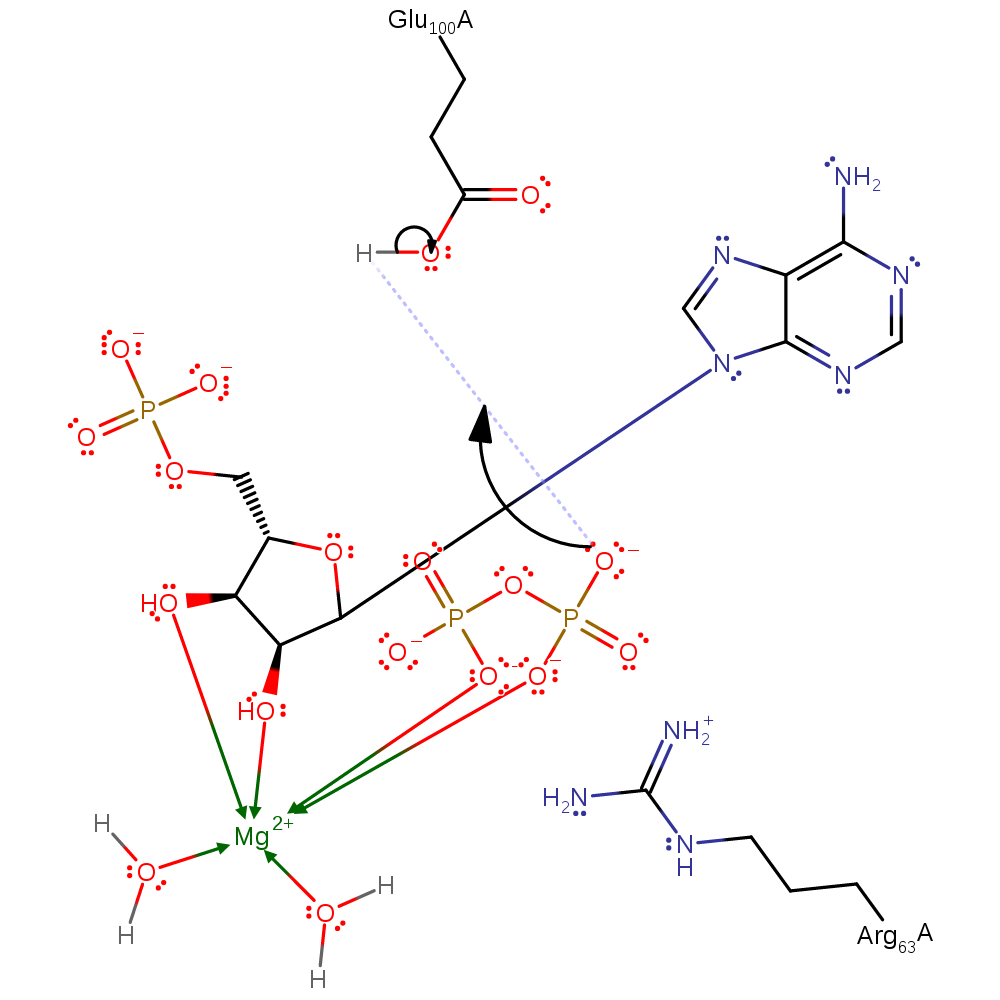
In the proposed Sn1 reaction, there is bond cleavage between the ribose and the pyrophosphate to form an oxocarbenium ion intermediate. This intermediate is subsequently attacked by N7 nucleophile on the adenine. Arg63 still stabilises the negatively charged pyrophosphate leaving group.
Catalytic Residues Roles
| UniProt | PDB* (1l1r) | ||
| Arg63 | Arg63A | Stabilises accumulation of negative charge on the pyrophosphate leaving group. | |
| Glu100 | Glu100A | Deprotonates N7 of adenine. | proton acceptor, proton donor |
Chemical Components
elimination (not covered by the Ingold mechanisms), heterolysis, intermediate formation, bimolecular nucleophilic addition, proton transfer, overall reactant used, overall product formed, inferred reaction step, native state of enzyme regeneratedReferences
- Arco JD et al. (2017), Curr Pharm Des,Purine and Pyrimidine Phosphoribosytransferases: A versatile tool for enzymatic synthesis of nucleoside-5'-monophosphates. DOI:10.2174/1381612823666171017165707. PMID:29046144.
- Takahashi R et al. (2010), J Biochem, 147, 95-107. Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase. DOI:10.1093/jb/mvp152. PMID:19819904.
- Silva CH et al. (2008), J Biomol Struct Dyn, 25, 589-597. Structural complexes of human adenine phosphoribosyltransferase reveal novel features of the APRT catalytic mechanism. DOI:10.1080/07391102.2008.10507205. PMID:18399692.
- Shi W et al. (2002), J Biol Chem, 277, 39981-39988. Closed Site Complexes of Adenine Phosphoribosyltransferase fromGiardia lamblia Reveal a Mechanism of Ribosyl Migration. DOI:10.1074/jbc.m205596200. PMID:12171925.
- Shi W et al. (2001), Biochemistry, 40, 10800-10809. Structural Analysis of Adenine Phosphoribosyltransferase fromSaccharomyces cerevisiae†,‡. DOI:10.1021/bi010465h. PMID:11535055.
Catalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
elimination (not covered by the Ingold mechanisms), heterolysis, intermediate formation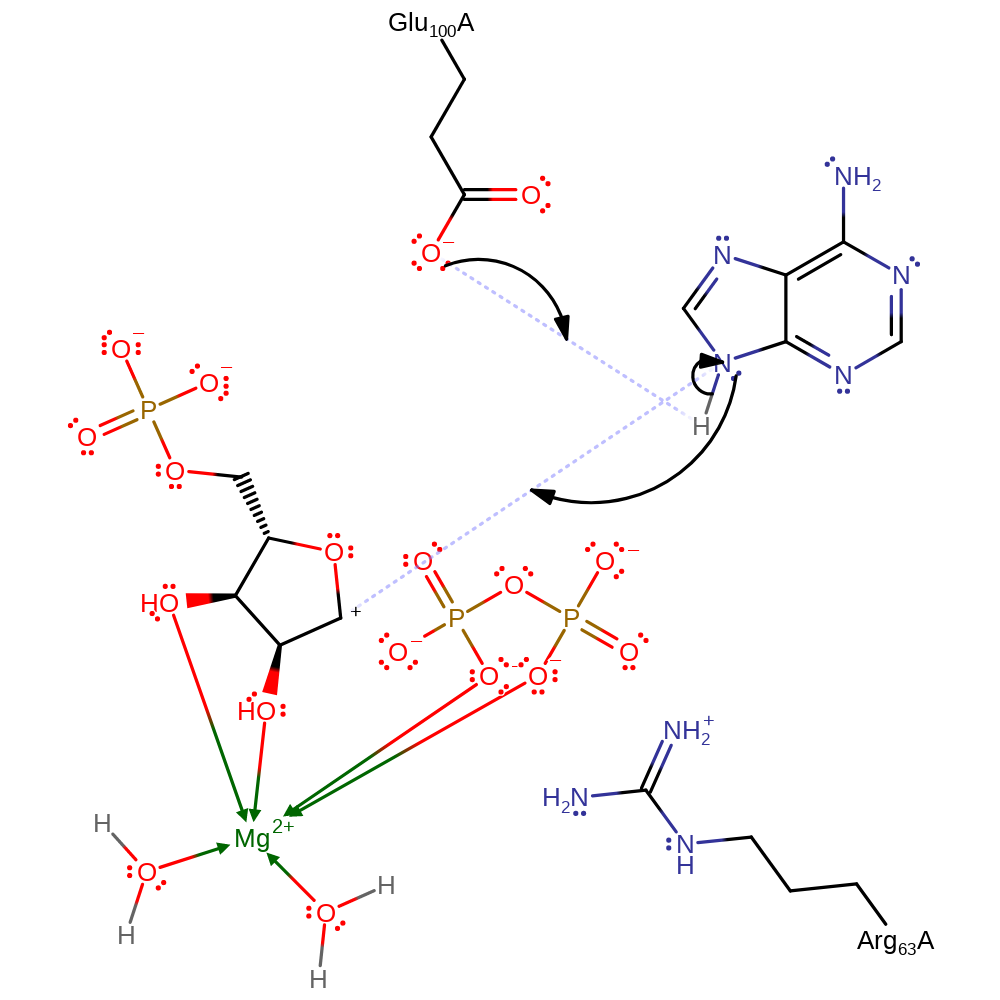
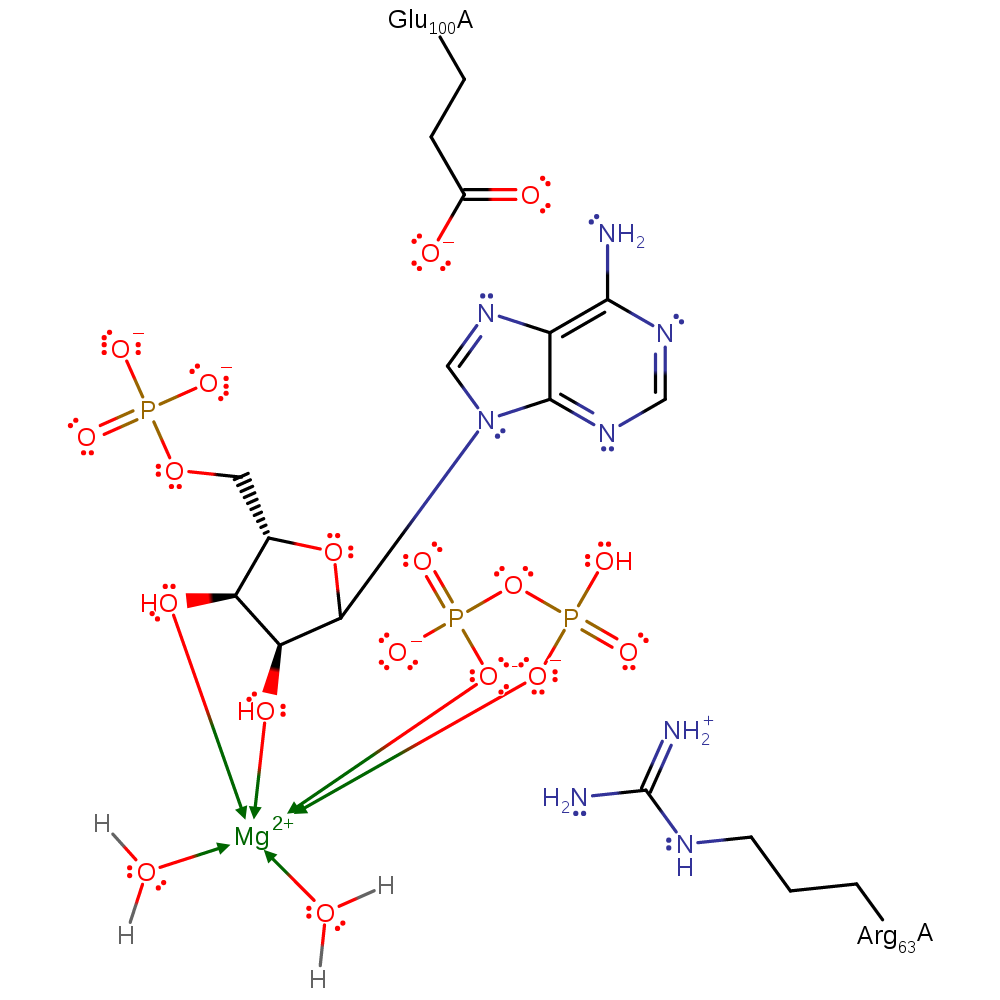
Step 2. Glu100 activates N7 by deprotonation to behave as a nucleophile and attack the oxocarbenium ion intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu100A | proton acceptor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, overall reactant usedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu100A | proton donor |
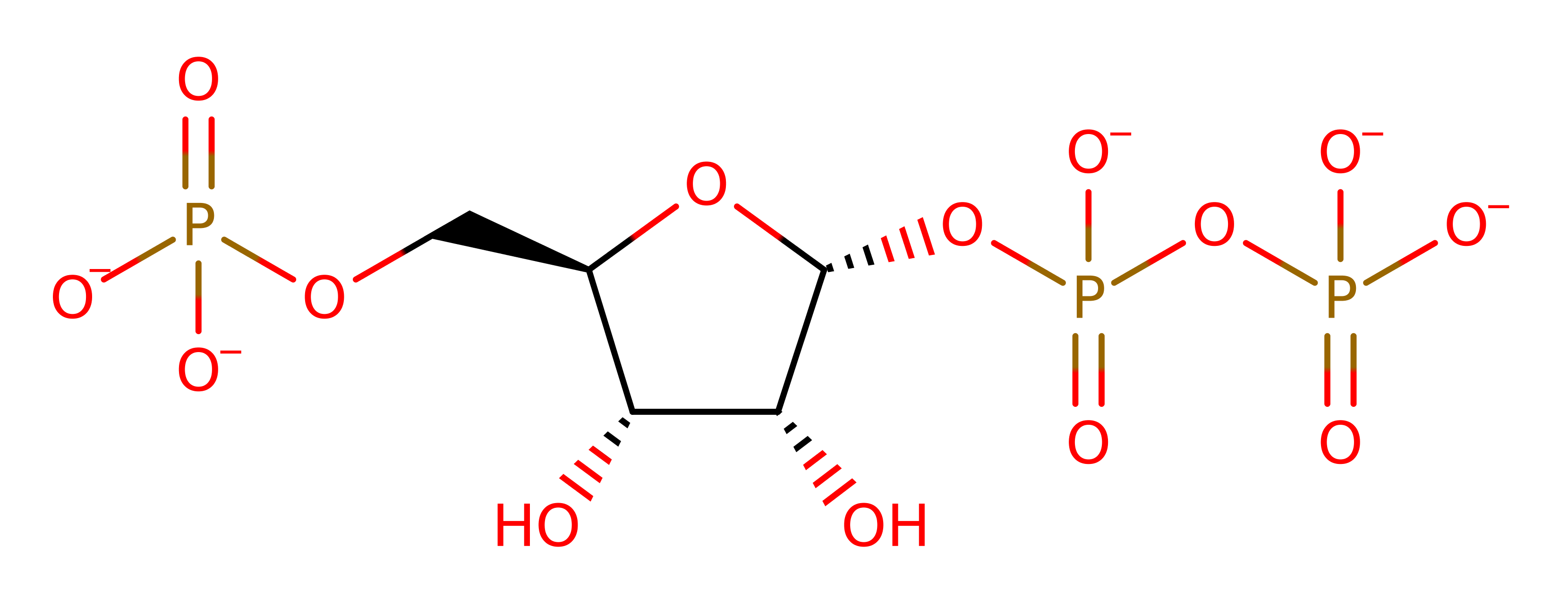



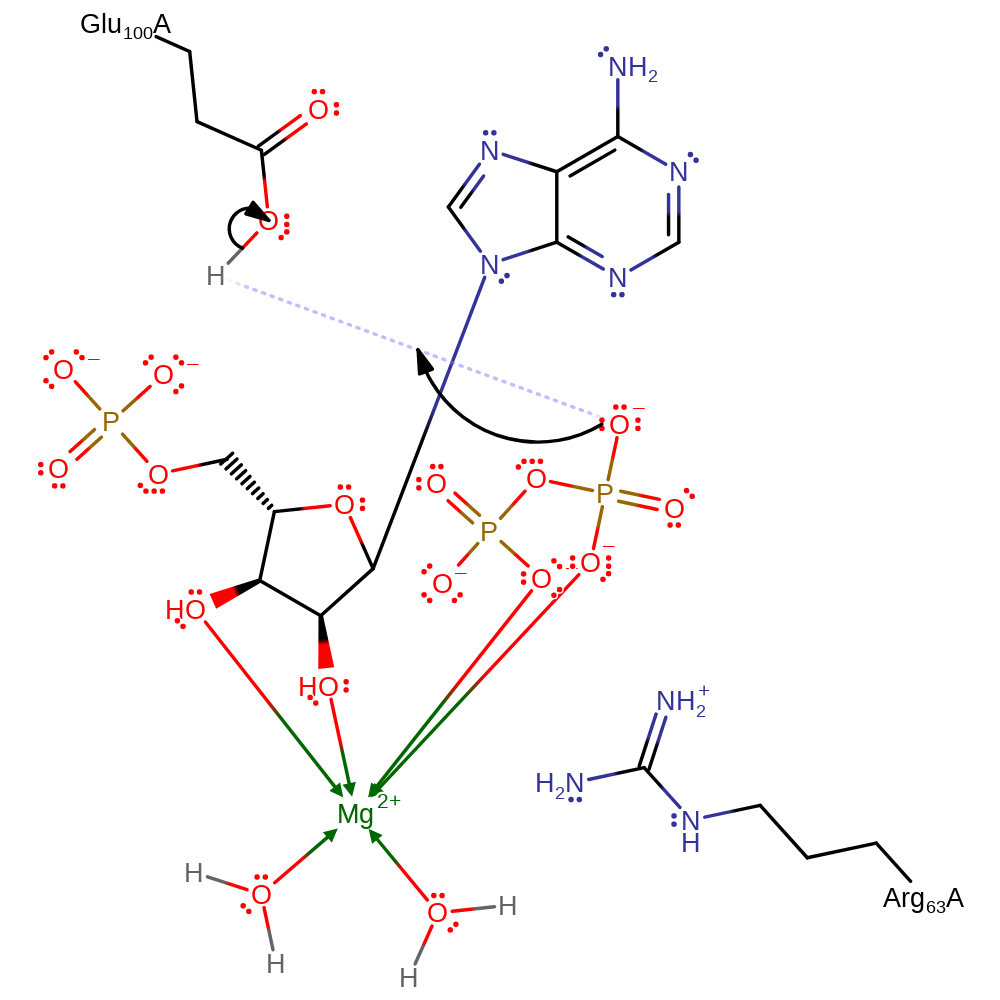
 Download:
Download: 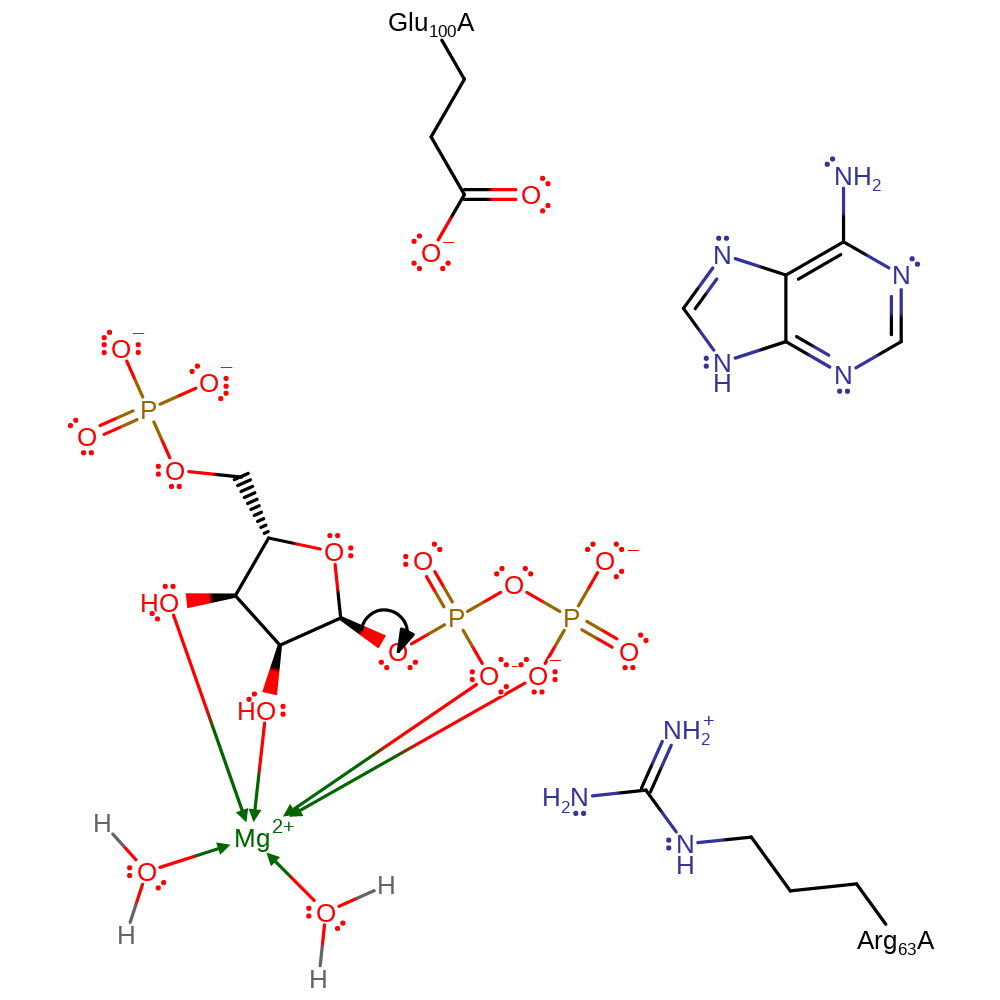
 Download:
Download: