Mitochondrial processing peptidase
Mitochondrial processing peptidase is a metalloendopeptidase that cleaves N-terminal signal sequences of proteins produced from nuclear information, transported from the cytosol to mitochondria. Cleavage of the signal sequence occurs at a single specific site. The enzyme shows functional and structural convergent evolution of this protease family with that of thermolysin, which has a very similar active site.
Reference Protein and Structure
- Sequences
-
P11914

P10507 (3.4.24.64)
(3.4.24.64)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Saccharomyces cerevisiae S288c (Baker's yeast)

- PDB
-
1hr6
- Yeast Mitochondrial Processing Peptidase
(2.5 Å)



- Catalytic CATH Domains
-
3.30.830.10
 (see all for 1hr6)
(see all for 1hr6)
- Cofactors
- Zinc(2+) (1)
Enzyme Reaction (EC:3.4.24.64)
Enzyme Mechanism
Introduction
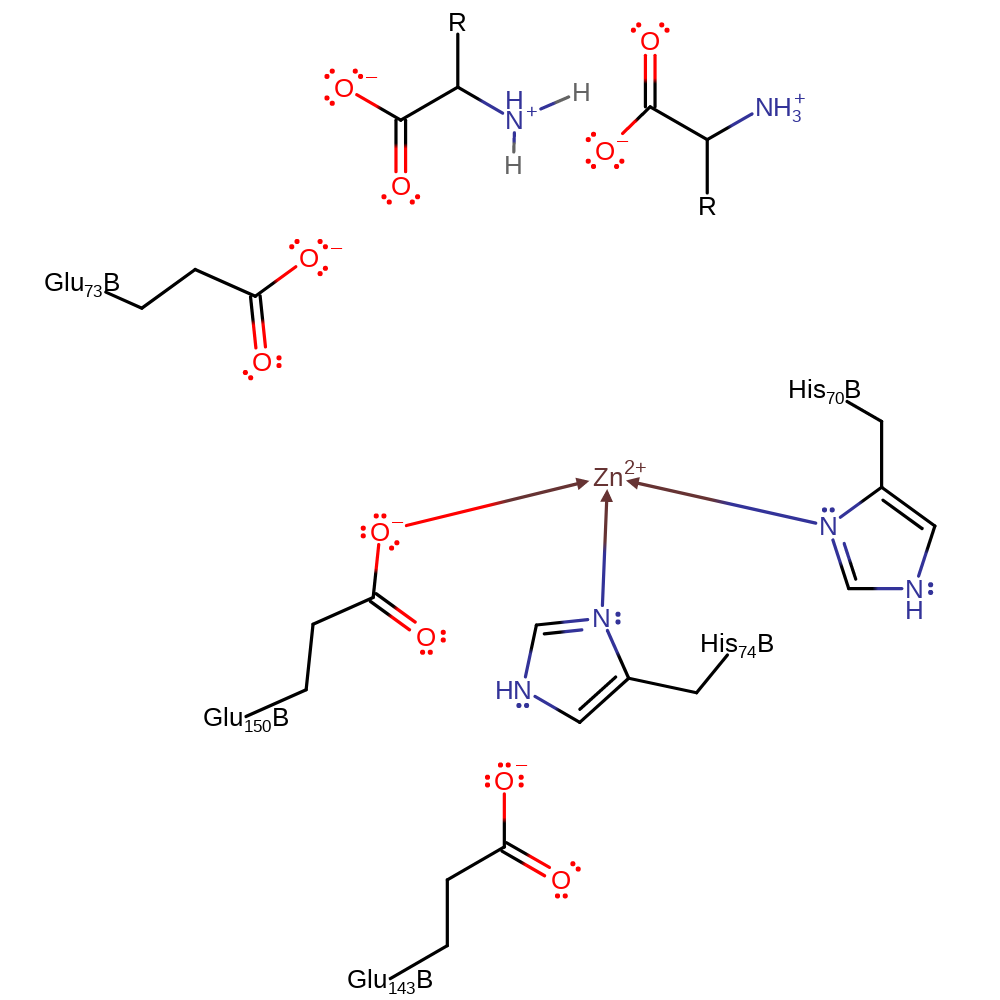
The mechanism of mitochondrial processing peptidase occurs in a thermolysin-like general-base-type mechanism. The zinc ion co-ordinates water which is displaced in substrate coordination towards Glu beta-73 which acts via general base catalysis to activate the water oxygen for nucleophilic attack on the carbonyl carbon of the scissile bond. The zinc ion also has a role in activating the water molecule by coordinating the oxygen. The pentacoordinate intermediate is formed and the proton accepted by Glu beta-73 in activating water is then transferred to the leaving nitrogen to facilitate collapse of the intermediate. A second proton transfer then occurs to shuttle a proton from the hydrated peptide to the leaving nitrogen.
Catalytic Residues Roles
| UniProt | PDB* (1hr6) | ||
| His70, His74, Glu150 | His70(51)B, His74(55)B, Glu150(131)B | Forms the Zinc binding site | metal ligand |
| Glu143 | Glu143(124)B | Forms a Hydrogen bond with His74 to stabilise it | hydrogen bond acceptor, electrostatic stabiliser |
| Glu73 | Glu73(54)B | Acts as a general acid/base catalyst in activating water for nucleophilic attack and collapse of intermediates through proton transfer to facilitate loss of the leaving group. | proton acceptor, proton donor |
Chemical Components
proton transfer, bimolecular nucleophilic addition, coordination to a metal ion, coordination, overall reactant used, intermediate formation, cofactor used, rate-determining step, unimolecular elimination by the conjugate base, heterolysis, intermediate collapse, decoordination from a metal ion, native state of cofactor regenerated, native state of enzyme regenerated, overall product formedReferences
- Taylor AB et al. (2001), Structure, 9, 615-625. Crystal Structures of Mitochondrial Processing Peptidase Reveal the Mode for Specific Cleavage of Import Signal Sequences. DOI:10.1016/s0969-2126(01)00621-9. PMID:11470436.
- Amata O et al. (2011), J Am Chem Soc, 133, 17824-17831. A proposal for mitochondrial processing peptidase catalytic mechanism. DOI:10.1021/ja207065v. PMID:21988451.
- Makarova KS et al. (1999), Protein Sci, 8, 2537-2540. Thermolysin and mitochondrial processing peptidase: How far structure-functional convergence goes. DOI:10.1110/ps.8.11.2537. PMID:10595562.
- Kitada S et al. (1995), J Biochem, 117, 1148-1150. A putative metal-binding site in the beta subunit of rat mitochondrial processing peptidase is essential for its catalytic activity. PMID:7490252.
- Becker AB et al. (1992), Proc Natl Acad Sci U S A, 89, 3835-3839. An unusual active site identified in a family of zinc metalloendopeptidases. DOI:10.1073/pnas.89.9.3835. PMID:1570301.
- Matthews BW (1988), Acc Chem Res, 21, 333-340. Structural basis of the action of thermolysin and related zinc peptidases. DOI:10.1021/ar00153a003.
- Hangauer DG et al. (1984), Biochemistry, 23, 5730-5741. An interactive computer graphics study of thermolysin-catalyzed peptide cleavage and inhibition by N-carboxymethyl dipeptides. DOI:10.1021/bi00319a011. PMID:6525336.

Step 1. Glu73B deprotonates water, which attacks the carbon of the carbonyl group in a nucleophilic addition. This results in coordination of the oxyanion to Zinc.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His70(51)B | metal ligand |
| His74(55)B | metal ligand |
| Glu150(131)B | metal ligand |
| Glu143(124)B | electrostatic stabiliser |
| Glu73(54)B | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, coordination to a metal ion, coordination, overall reactant used, intermediate formation, cofactor used, rate-determining step
Step 2. The oxyanion initiates an elimination which results in the cleavage of the peptide bond. The N-terminal product then accepts a proton from Glu73B.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His70(51)B | metal ligand |
| His74(55)B | metal ligand |
| Glu150(131)B | metal ligand |
| Glu143(124)B | electrostatic stabiliser, hydrogen bond acceptor |
| His74(55)B | hydrogen bond donor |
| Glu73(54)B | proton donor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, heterolysis, intermediate collapse, decoordination from a metal ion, native state of cofactor regenerated, native state of enzyme regenerated
Step 3. The N-terminal product deprotonates the C-terminal product to form kinetically favourable products.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His70(51)B | metal ligand |
| His74(55)B | metal ligand, hydrogen bond donor |
| Glu143(124)B | hydrogen bond acceptor, electrostatic stabiliser |
| Glu150(131)B | metal ligand |



 Download:
Download: