Methylmalonyl-CoA mutase
Isomerising two important metabolites gives this enzyme significance in the degradation of some amino acids and odd-chain fatty acids, as well as (in bacteria) the biosynthesis of propionate. Like several other mutases, it uses adenosylcobalamin (Vitamin B12) to produce the free radicals essential for the rearrangement, and its sequence is rather well conserved between prokaryotes and eukaryotes.
Reference Protein and Structure
- Sequences
-
P11653
 (5.4.99.2)
(5.4.99.2)
P11652 (5.4.99.2)
(5.4.99.2)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Propionibacterium freudenreichii subsp. shermanii (Bacteria)

- PDB
-
1req
- METHYLMALONYL-COA MUTASE
(2.0 Å)



- Catalytic CATH Domains
-
3.40.50.280
 3.20.20.240
3.20.20.240  (see all for 1req)
(see all for 1req)
- Cofactors
- Cob(iii)alamin (1) Metal MACiE
Enzyme Reaction (EC:5.4.99.2)
Enzyme Mechanism
Introduction
The protein can assist in catalysis in two ways; firstly by encouraging the homolytic cleavage of the Co-C bond in the cofactor, and then during the rearrangement itself. In the first of these two considerations, the axial ligand His610 is clearly important - the 2.5 A bond it forms to Co encourages formation of the active Co(II) species. To maintain this bond length, Asp608 forms a strong H-bond to His610, but this will induce a negative charge on the His, strengthening the Co-C bond. This undesired side-effect is compensated for by Lys604, which is positively charged and bonds to the other side of Asp608. In addition to this triad of residues, several are involved in the rearrangement, although a detailed catalytic mechanism has not yet been elucidated. Arg207 seems to be important in holding the carboxyl group of the substrate in the correct position for catalysis. His244 stabilises the intermediate, as shown by mutagenesis. Mutagenesis also demonstrates that changing Tyr89 to Phe slows both the homolysis of the Co-C bond and the rearrangement itself.
Catalytic Residues Roles
| UniProt | PDB* (1req) | ||
| His244 | His244(243)A | Stabilises the reactive intermediates. | proton acceptor, electrostatic stabiliser, proton donor |
| His610 | His610(609)A | Forms the axial ligand of the cobalt of the cobalamin cofactor. | metal ligand |
| Tyr243 | Tyr243(242)A | This residue helps to minimise radical extinction side reactions. | radical stabiliser, electrostatic stabiliser |
| Tyr89 | Tyr89(88)A | Helps stabilise the reactive radical intermediates. | radical stabiliser, electrostatic stabiliser |
| Asp608, Lys604 | Asp608(607)A, Lys604(603)A | Important in maintaining the His610-Co bond length. Asp608 forms a strong H-bond to His610, but this will induce a negative charge on the His, strengthening the Co-C bond. This undesired side-effect is compensated for by Lys604, which is positively charged and bonds to the other side of Asp608. | electrostatic stabiliser |
Chemical Components
radical formation, homolysis, cofactor used, coordination to a metal ion, decoordination from a metal ion, intermediate formation, radical propagation, hydrogen transfer, overall reactant used, proton transfer, intramolecular homolytic addition, cyclisation, bimolecular homolytic elimination, decyclisation, overall product formed, intermediate terminated, colligation, radical termination, native state of enzyme regenerated, native state of cofactor regeneratedReferences
- Mancia F et al. (1999), Biochemistry, 38, 7999-8005. Crystal Structure of Substrate Complexes of Methylmalonyl-CoA Mutase. DOI:10.1021/bi9903852. PMID:10387043.
- Makins C et al. (2013), Biochemistry, 52, 878-888. Mutagenesis of a Conserved Glutamate Reveals the Contribution of Electrostatic Energy to Adenosylcobalamin Co–C Bond Homolysis in Ornithine 4,5-Aminomutase and Methylmalonyl-CoA Mutase. DOI:10.1021/bi3012719. PMID:23311430.
- Takahashi-Iñiguez T et al. (2012), J Zhejiang Univ Sci B, 13, 423-437. Role of vitamin B12 on methylmalonyl-CoA mutase activity. DOI:10.1631/jzus.b1100329. PMID:22661206.
- Kumar N et al. (2012), J Phys Chem Lett, 3, 1035-1038. Charge Separation Propensity of the Coenzyme B12–Tyrosine Complex in Adenosylcobalamin-Dependent Methylmalonyl–CoA Mutase Enzyme. DOI:10.1021/jz300102s. PMID:26286568.
- Dybala-Defratyka A et al. (2007), Proc Natl Acad Sci U S A, 104, 10774-10779. Coupling of hydrogenic tunneling to active-site motion in the hydrogen radical transfer catalyzed by a coenzyme B12-dependent mutase. DOI:10.1073/pnas.0702188104. PMID:17581872.
- Banerjee R et al. (2006), Philos Trans R Soc Lond B Biol Sci, 361, 1333-1339. Quantum catalysis in B12-dependent methylmalonyl-CoA mutase: experimental and computational insights. DOI:10.1098/rstb.2006.1866. PMID:16873121.
- Padovani D et al. (2006), Biochemistry, 45, 2951-2959. Alternative Pathways for Radical Dissipation in an Active Site Mutant of B12-Dependent Methylmalonyl-CoA Mutase†. DOI:10.1021/bi051742d. PMID:16503649.
- Mansoorabadi SO et al. (2005), Biochemistry, 44, 3153-3158. Characterization of a Succinyl-CoA Radical−Cob(II)alamin Spin Triplet Intermediate in the Reaction Catalyzed by Adenosylcobalamin-Dependent Methylmalonyl-CoA Mutase†. DOI:10.1021/bi0482102. PMID:15736925.
- Marsh EN et al. (2001), Curr Opin Chem Biol, 5, 499-505. Adenosylcobalamin-dependent isomerases: new insights into structure and mechanism. DOI:10.1016/s1367-5931(00)00238-6. PMID:11578922.
- Smith DM et al. (2001), J Am Chem Soc, 123, 1664-1675. Understanding the Mechanism of B12-Dependent Diol Dehydratase: A Synergistic Retro-Push−Pull Proposal. DOI:10.1021/ja001454z. PMID:11456766.
- Wetmore SD et al. (2001), Chembiochem, 2, 919-922. Catalysis by mutants of methylmalonyl-CoA mutase: a theoretical rationalization for a change in the rate-determining step. PMID:11948881.
- Thomä NH et al. (2000), Biochemistry, 39, 9213-9221. Protection of Radical Intermediates at the Active Site of Adenosylcobalamin-Dependent Methylmalonyl-CoA Mutase†,‡. DOI:10.1021/bi0004302. PMID:10924114.
- Thomä NH et al. (1998), Biochemistry, 37, 14386-14393. Stabilization of Radical Intermediates by an Active-Site Tyrosine Residue in Methylmalonyl-CoA Mutase†,‡. DOI:10.1021/bi981375o. PMID:9772164.
- Mancia F et al. (1998), Structure, 6, 711-720. Conformational changes on substrate binding to methylmalonyl CoA mutase and new insights into the free radical mechanism. PMID:9655823.
- Mancia F et al. (1996), Structure, 4, 339-350. How coenzyme B12 radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2 å resolution. DOI:10.1016/s0969-2126(96)00037-8. PMID:8805541.
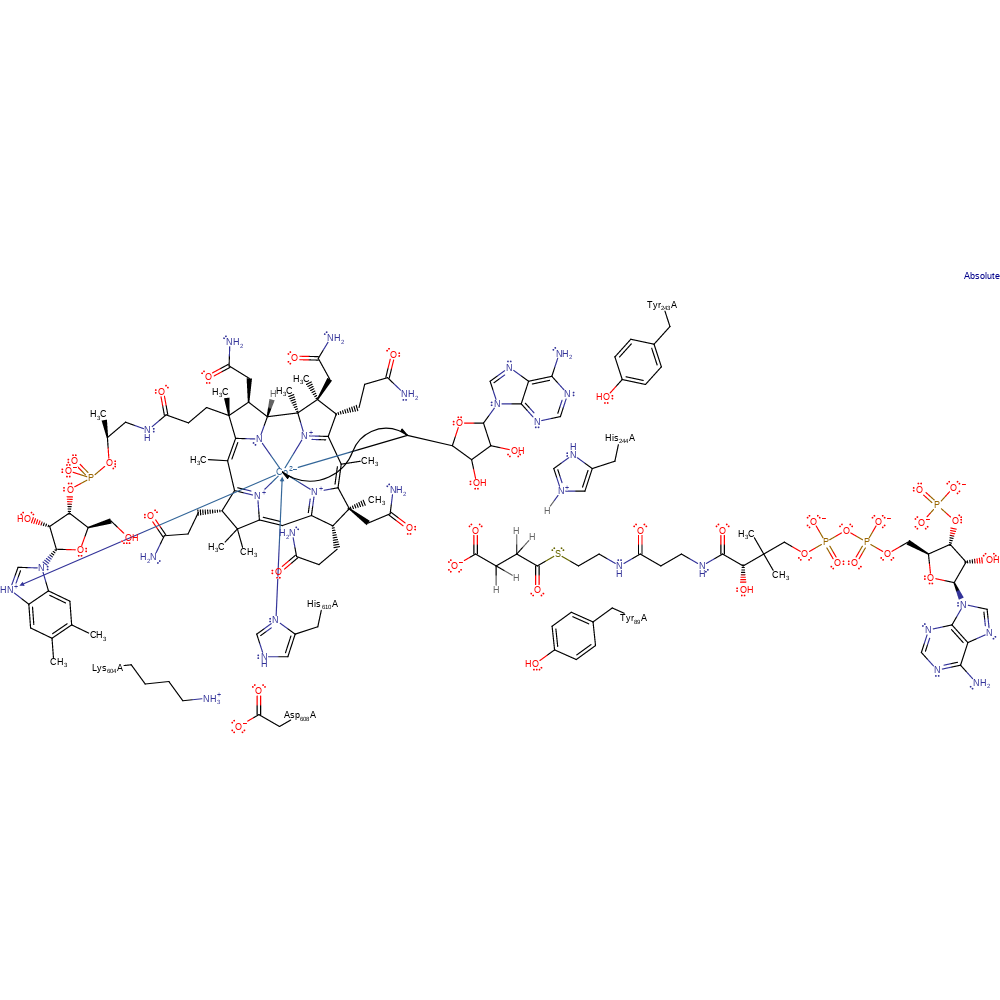
Step 1. The Co(II)-C bond of the B12 cofactor undergoes homoloysis, forming the adenosyl radical and Co(I).
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His610(609)A | metal ligand |
| Lys604(603)A | electrostatic stabiliser |
| Asp608(607)A | electrostatic stabiliser |
Chemical Components
radical formation, homolysis, cofactor used, coordination to a metal ion, decoordination from a metal ion, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr89(88)A | radical stabiliser |
| Tyr243(242)A | radical stabiliser |
| His610(609)A | metal ligand |
| His244(243)A | electrostatic stabiliser |
| Lys604(603)A | electrostatic stabiliser |
| Asp608(607)A | electrostatic stabiliser |
Chemical Components
radical propagation, hydrogen transfer, overall reactant used, intermediate formation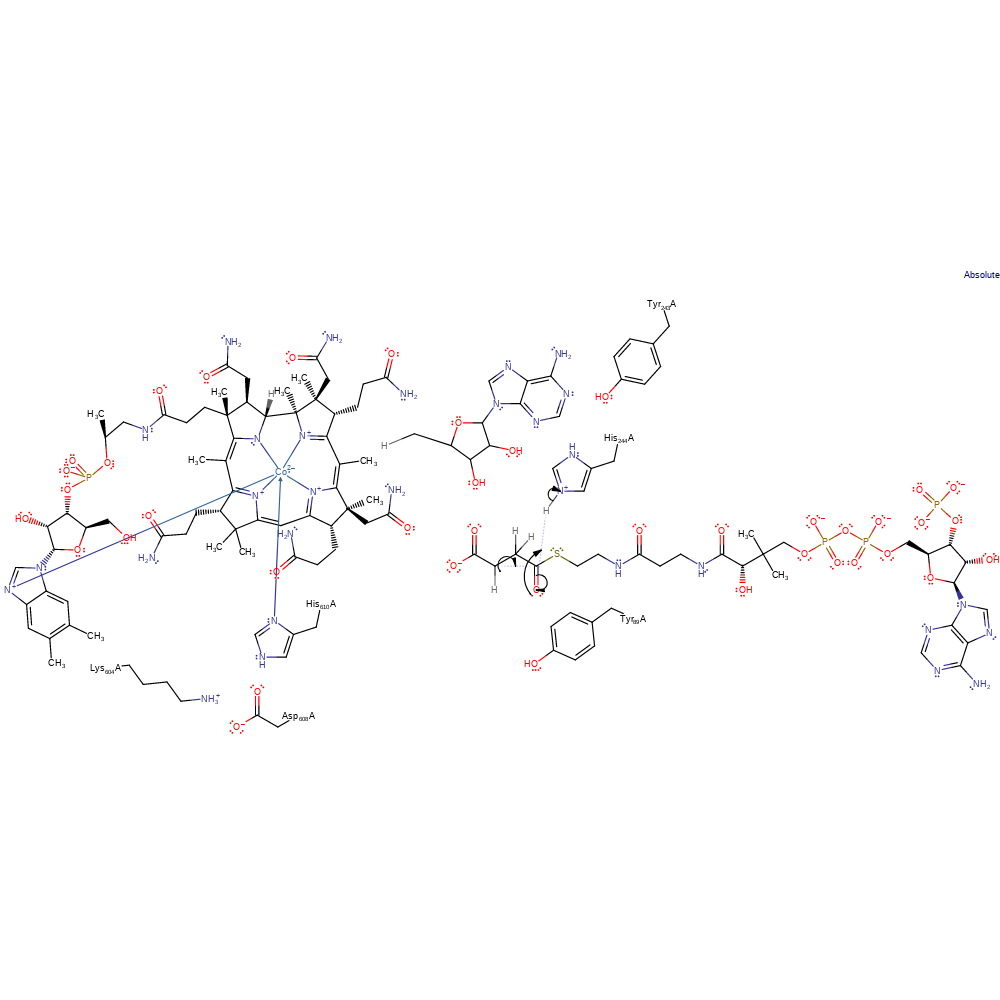
Step 3. The substrate radical attacks its own carbonyl carbon, forcing the radical onto the oxygen, which deprotonates His244.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr89(88)A | electrostatic stabiliser |
| Tyr243(242)A | electrostatic stabiliser |
| His610(609)A | metal ligand |
| Lys604(603)A | electrostatic stabiliser |
| Asp608(607)A | electrostatic stabiliser |
| His244(243)A | proton donor |
Chemical Components
proton transfer, radical propagation, ingold: intramolecular homolytic addition, cyclisation, intermediate formation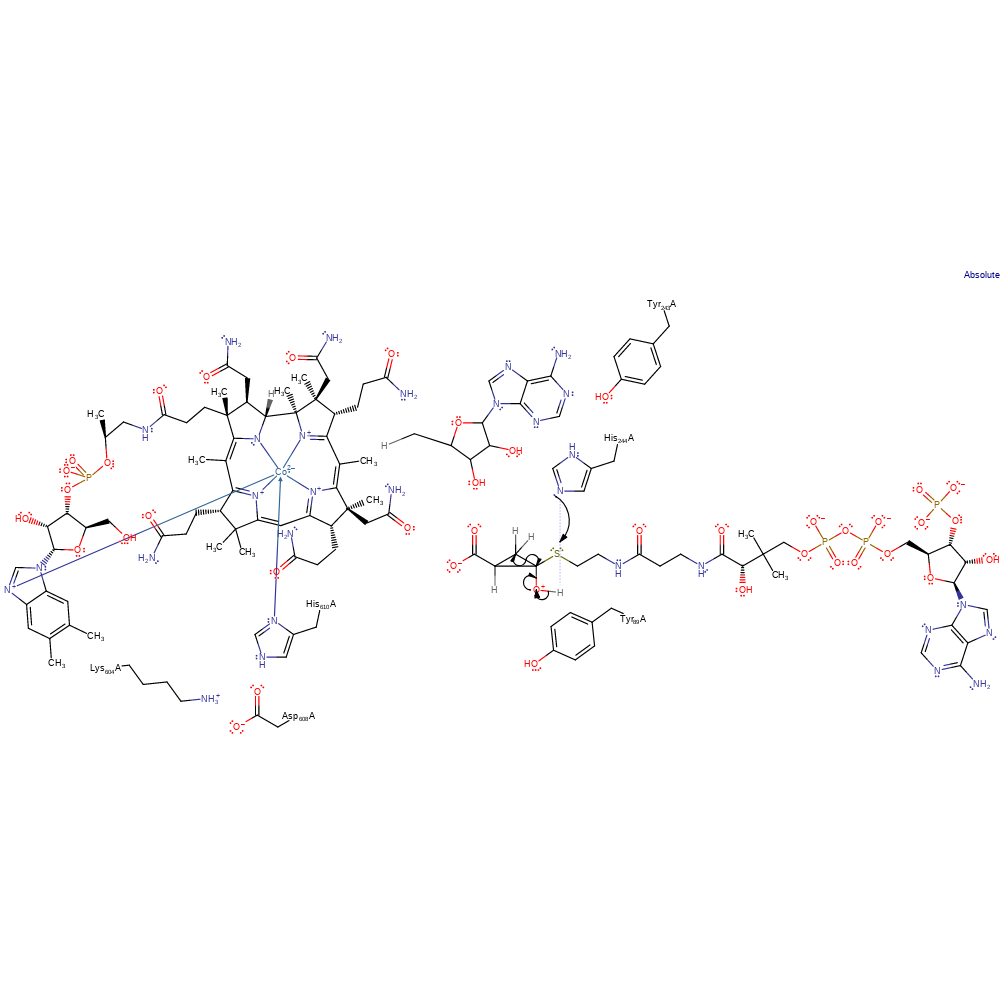
Step 4. His244 deprotonates the oxygen, which causes radical rearrangement and the cleavage of the propane ring formed, the radical ends up on the newly formed CH2 branch.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr89(88)A | radical stabiliser |
| Tyr243(242)A | radical stabiliser |
| Lys604(603)A | electrostatic stabiliser |
| Asp608(607)A | electrostatic stabiliser |
| His610(609)A | metal ligand |
| His244(243)A | proton acceptor |
Chemical Components
radical propagation, ingold: bimolecular homolytic elimination, decyclisation, intermediate formation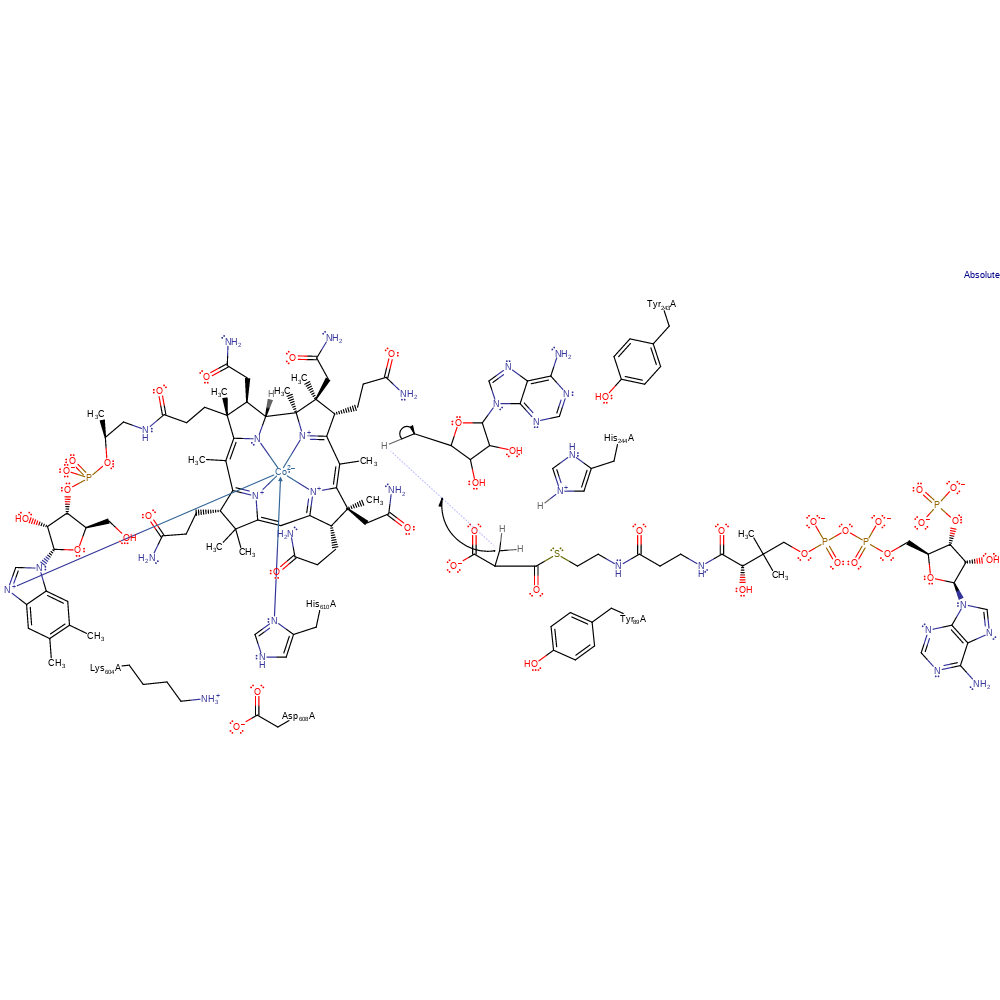
Step 5. The substrate radical abstracts a hydrogen from the adenosine, re-forming the adenosyl radical.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr89(88)A | radical stabiliser |
| Tyr243(242)A | radical stabiliser |
| Lys604(603)A | electrostatic stabiliser |
| Asp608(607)A | electrostatic stabiliser |
| His610(609)A | metal ligand |
Chemical Components
hydrogen transfer, radical propagation, overall product formed, intermediate terminated, intermediate formation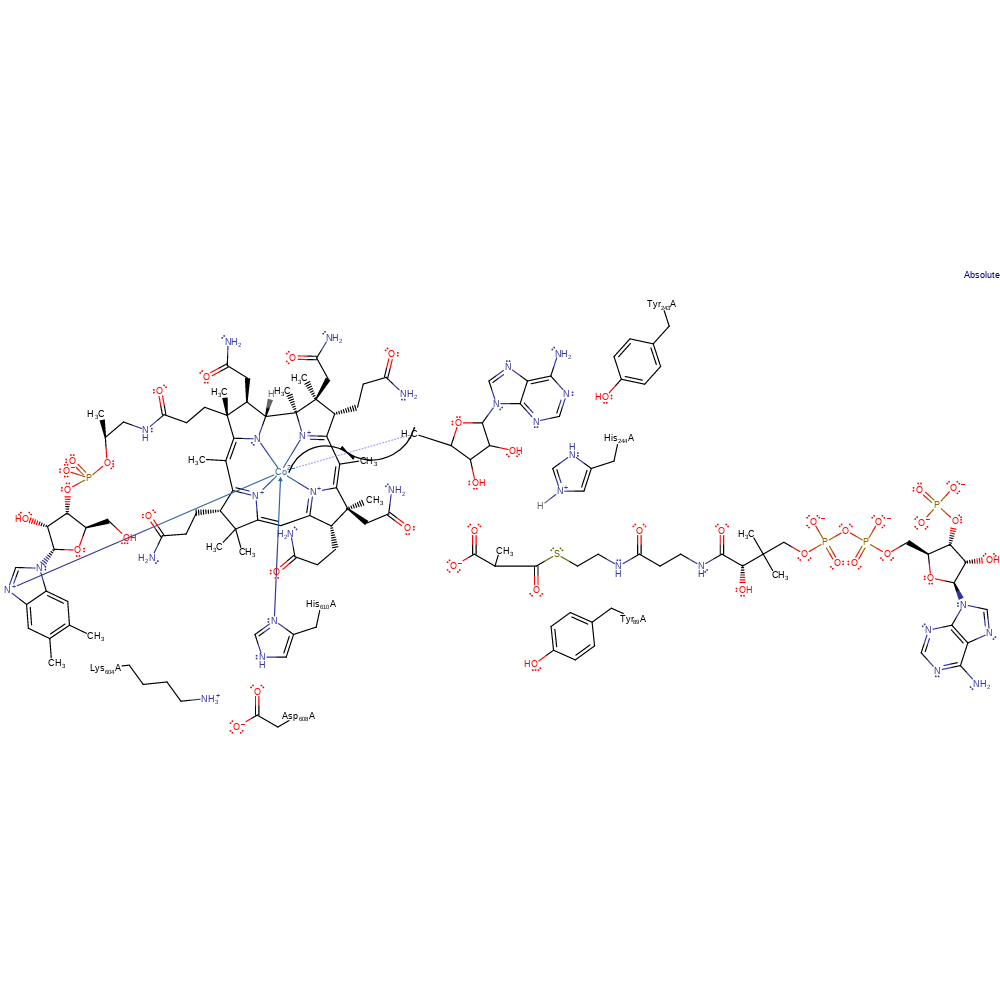
Step 6. The adenosyl radical and Co(I) undergo colligation, reforming the B12 cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys604(603)A | electrostatic stabiliser |
| Asp608(607)A | electrostatic stabiliser |
| His610(609)A | metal ligand |


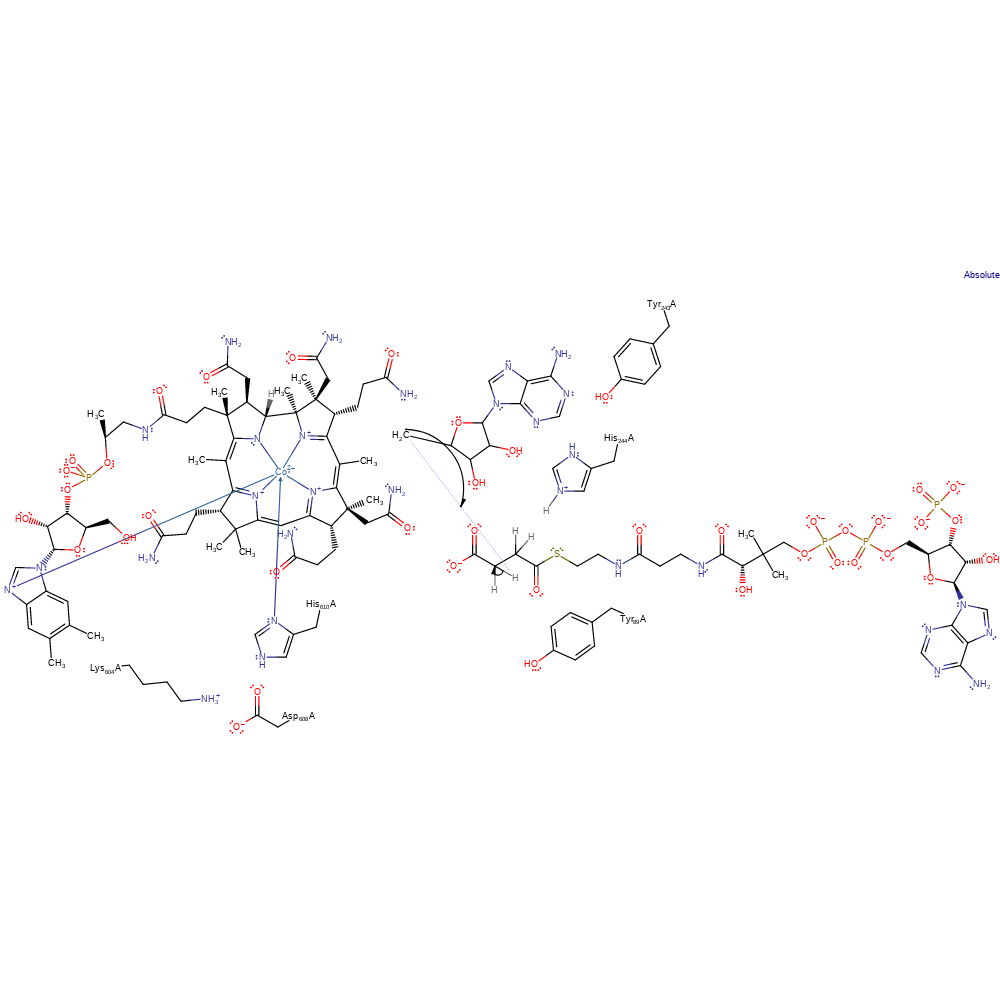
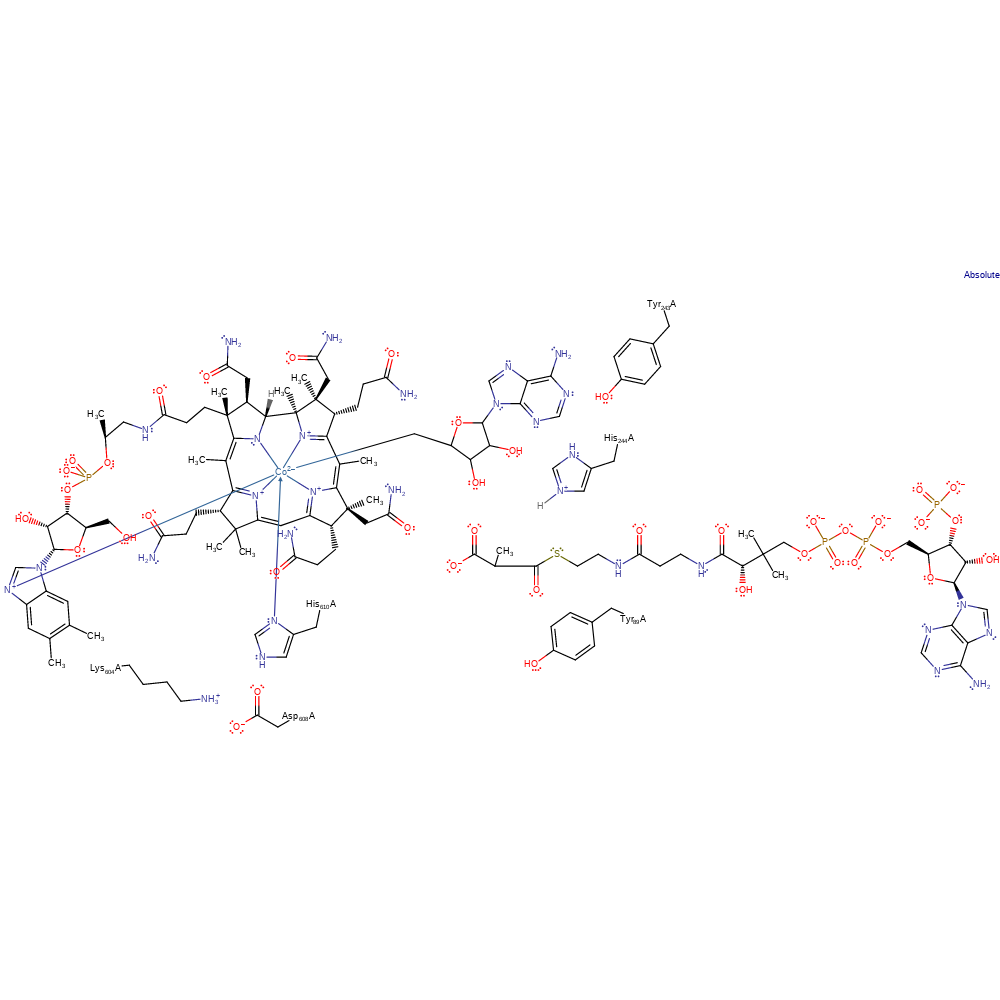 Download:
Download: