
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D:
E.C.5.4.99.2
- methylmalonyl-CoA mutase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(R)-methylmalonyl-CoA = succinyl-CoA
|
 |
 |
 |
 |
 |
(R)-methylmalonyl-CoA
Bound ligand (Het Group name = )
matches with 85.45% similarity
|
=
|
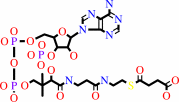 succinyl-CoA
succinyl-CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Cob(II)alamin
|
 |
 |
 |
 |
 |
Cob(II)alamin
Bound ligand (Het Group name =
B12)
matches with 85.71% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
4:339-350
(1996)
|
|
PubMed id:
|
|
|
|

|
| |
|
How coenzyme B12 radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2 A resolution.
|
|
F.Mancia,
N.H.Keep,
A.Nakagawa,
P.F.Leadlay,
S.McSweeney,
B.Rasmussen,
P.Bösecke,
O.Diat,
P.R.Evans.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: The enzyme methylmalonyl-coenzyme A (CoA) mutase, an alphabeta
heterodimer of 150 kDa, is a member of a class of enzymes that uses coenzyme B12
(adenosylcobalamin) as a cofactor. The enzyme induces the formation of an
adenosyl radical from the cofactor. This radical then initiates a free-radical
rearrangement of its substrate, succinyl-CoA, to methylmalonyl-CoA. RESULTS:
Reported here is the crystal structure at 2 A resolution of methylmalonyl-CoA
mutase from Propionibacterium shermanii in complex with coenzyme B12 and with
the partial substrate desulpho-CoA (lacking the succinyl group and the sulphur
atom of the substrate). The coenzyme is bound by a domain which shares a similar
fold to those of flavodoxin and the B12-binding domain of
methylcobalamin-dependent methionine synthase. The cobalt atom is coordinated,
via a long bond, to a histidine from the protein. The partial substrate is bound
along the axis of a (beta/alpha)8 TIM barrel domain. CONCLUSIONS: The
histidine-cobalt distance is very long (2.5 A compared with 1.95-2.2 A in free
cobalamins), suggesting that the enzyme positions the histidine in order to
weaken the metal-carbon bond of the cofactor and favour the formation of the
initial radical species. The active site is deeply buried, and the only access
to it is through a narrow tunnel along the axis of the TIM barrel domain.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. Part of the electron density map at 3 å
resolution that was used to build the model, showing the β
sheet of the C-terminal domain of the α chain, with the refined
model superimposed. Figure 2. Part of the electron density
map at 3 å resolution that was used to build the model,
showing the β sheet of the C-terminal domain of the α chain,
with the refined model superimposed.
|
 |
Figure 7.
Figure 7. Schematic representation of ligand binding. (a) The
interactions between protein and coenzyme B[12]. The hydrophobic
pocket for the dimethylbenzimidazole is lined by residues
IleA617, TyrA705, GlyA685 and SerA655: this last forms a
hydrogen bond to the N3B nitrogen atom of the base, which is the
atom that coordinates the cobalt atom in the free coenzyme.
LeuA657 stacks against HisA610, the residue that coordinates the
cobalt, and forms hydrophobic interactions with the beginning
of the pseudo-nucleotide tail (C56) and with the C20 methyl
group of the corrin. (b) The interactions between protein and
desulpho-CoA. TyrA75 stacks on the adenine ring. Note that
ArgB45 is the only interaction between the β chain and the
substrate. CoA would have an additional thiol group attached to
the left-hand end. Figure 7. Schematic representation of
ligand binding. (a) The interactions between protein and
coenzyme B[12]. The hydrophobic pocket for the
dimethylbenzimidazole is lined by residues IleA617, TyrA705,
GlyA685 and SerA655: this last forms a hydrogen bond to the N3B
nitrogen atom of the base, which is the atom that coordinates
the cobalt atom in the free coenzyme. LeuA657 stacks against
HisA610, the residue that coordinates the cobalt, and forms
hydrophobic interactions with the beginning of the
pseudo-nucleotide tail (C56) and with the C20 methyl group of
the corrin. (b) The interactions between protein and
desulpho-CoA. TyrA75 stacks on the adenine ring. Note that
ArgB45 is the only interaction between the β chain and the
substrate. CoA would have an additional thiol group attached to
the left-hand end.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Structure
(1996,
4,
339-350)
copyright 1996.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.S.Froese,
and
R.A.Gravel
(2010).
Genetic disorders of vitamin B₁₂ metabolism: eight complementation groups--eight genes.
|
| |
Expert Rev Mol Med,
12,
e37.
|
 |
|
|
|
|
 |
E.N.Marsh,
D.P.Patterson,
and
L.Li
(2010).
Adenosyl radical: reagent and catalyst in enzyme reactions.
|
| |
Chembiochem,
11,
604-621.
|
 |
|
|
|
|
 |
P.E.Mera,
and
J.C.Escalante-Semerena
(2010).
Multiple roles of ATP:cob(I)alamin adenosyltransferases in the conversion of B12 to coenzyme B12.
|
| |
Appl Microbiol Biotechnol,
88,
41-48.
|
 |
|
|
|
|
 |
T.Toraya
(2010).
[Microbe-inspired system enzymology of vitamin B₁₂ metabolism].
|
| |
Yakugaku Zasshi,
130,
1453-1462.
|
 |
|
|
|
|
 |
B.M.Alzoubi,
F.Vidali,
R.Puchta,
C.Dücker-Benfer,
A.Felluga,
L.Randaccio,
G.Tauzher,
and
R.van Eldik
(2009).
Mechanistic behaviour of alkylcobaloximes and imino-oxime complexes related to vitamin B(12).
|
| |
Dalton Trans,
(),
2392-2399.
|
 |
|
|
|
|
 |
M.J.Gray,
and
J.C.Escalante-Semerena
(2009).
The cobinamide amidohydrolase (cobyric acid-forming) CbiZ enzyme: a critical activity of the cobamide remodelling system of Rhodobacter sphaeroides.
|
| |
Mol Microbiol,
74,
1198-1210.
|
 |
|
|
|
|
 |
N.Sukumar,
F.S.Mathews,
M.M.Gordon,
S.E.Ealick,
and
D.H.Alpers
(2009).
Postcrystallization Analysis of the Irreproducibility of the Human Intrinsic Factor-Cobalamin Complex Crystals.
|
| |
Cryst Growth Des,
9,
348-351.
|
 |
|
|
|
|
 |
P.E.Mera,
M.St Maurice,
I.Rayment,
and
J.C.Escalante-Semerena
(2009).
Residue Phe112 of the human-type corrinoid adenosyltransferase (PduO) enzyme of Lactobacillus reuteri is critical to the formation of the four-coordinate Co(II) corrinoid substrate and to the activity of the enzyme.
|
| |
Biochemistry,
48,
3138-3145.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Banerjee,
C.Gherasim,
and
D.Padovani
(2009).
The tinker, tailor, soldier in intracellular B12 trafficking.
|
| |
Curr Opin Chem Biol,
13,
484-491.
|
 |
|
|
|
|
 |
A.Chatterjee,
Y.Li,
Y.Zhang,
T.L.Grove,
M.Lee,
C.Krebs,
S.J.Booker,
T.P.Begley,
and
S.E.Ealick
(2008).
Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily.
|
| |
Nat Chem Biol,
4,
758-765.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Kim,
C.Gherasim,
and
R.Banerjee
(2008).
Decyanation of vitamin B12 by a trafficking chaperone.
|
| |
Proc Natl Acad Sci U S A,
105,
14551-14554.
|
 |
|
|
|
|
 |
S.Gallo,
M.Oberhuber,
R.K.Sigel,
and
B.Kräutler
(2008).
The corrin moiety of coenzyme B12 is the determinant for switching the btuB riboswitch of E. coli.
|
| |
Chembiochem,
9,
1408-1414.
|
 |
|
|
|
|
 |
S.Savvi,
D.F.Warner,
B.D.Kana,
J.D.McKinney,
V.Mizrahi,
and
S.S.Dawes
(2008).
Functional characterization of a vitamin B12-dependent methylmalonyl pathway in Mycobacterium tuberculosis: implications for propionate metabolism during growth on fatty acids.
|
| |
J Bacteriol,
190,
3886-3895.
|
 |
|
|
|
|
 |
T.Toraya,
N.Tamura,
T.Watanabe,
M.Yamanishi,
N.Hieda,
and
K.Mori
(2008).
Mechanism-based inactivation of coenzyme B12-dependent diol dehydratase by 3-unsaturated 1,2-diols and thioglycerol.
|
| |
J Biochem,
144,
437-446.
|
 |
|
|
|
|
 |
C.Oyama,
T.Takahashi,
M.Matsumori,
Y.Shoji,
G.Tajima,
N.Sakura,
Y.Hasegawa,
S.Yamaguchi,
H.Kakinuma,
and
G.Takada
(2007).
Novel mutation of methylmalonyl-CoA mutase gene causing the mut0 form of methylmalonic acidemia in a Japanese girl.
|
| |
Pediatr Int,
49,
232-234.
|
 |
|
|
|
|
 |
C.Wheatley
(2007).
The return of the Scarlet Pimpernel: cobalamin in inflammation II - cobalamins can both selectively promote all three nitric oxide synthases (NOS), particularly iNOS and eNOS, and, as needed, selectively inhibit iNOS and nNOS.
|
| |
J Nutr Environ Med,
16,
181-211.
|
 |
|
|
|
|
 |
C.Wheatley
(2007).
Cobalamin in inflammation III - glutathionylcobalamin and methylcobalamin/adenosylcobalamin coenzymes: the sword in the stone? How cobalamin may directly regulate the nitric oxide synthases.
|
| |
J Nutr Environ Med,
16,
212-226.
|
 |
|
|
|
|
 |
L.Hannibal,
S.D.Bunge,
R.van Eldik,
D.W.Jacobsen,
C.Kratky,
K.Gruber,
and
N.E.Brasch
(2007).
X-ray structural characterization of imidazolylcobalamin and histidinylcobalamin: cobalamin models for aquacobalamin bound to the B12 transporter protein transcobalamin.
|
| |
Inorg Chem,
46,
3613-3618.
|
 |
|
|
|
|
 |
P.M.Kozlowski,
T.Kamachi,
T.Toraya,
and
K.Yoshizawa
(2007).
Does Cob(II)alamin act as a conductor in coenzyme B12 dependent mutases?
|
| |
Angew Chem Int Ed Engl,
46,
980-983.
|
 |
|
|
|
|
 |
S.Watanabe,
R.Matsumi,
T.Arai,
H.Atomi,
T.Imanaka,
and
K.Miki
(2007).
Crystal structures of [NiFe] hydrogenase maturation proteins HypC, HypD, and HypE: insights into cyanation reaction by thiol redox signaling.
|
| |
Mol Cell,
27,
29-40.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.L.Brown
(2006).
The enzymatic activation of coenzyme B12.
|
| |
Dalton Trans,
(),
1123-1133.
|
 |
|
|
|
|
 |
L.C.Worgan,
K.Niles,
J.C.Tirone,
A.Hofmann,
A.Verner,
A.Sammak,
T.Kucic,
P.Lepage,
and
D.S.Rosenblatt
(2006).
Spectrum of mutations in mut methylmalonic acidemia and identification of a common Hispanic mutation and haplotype.
|
| |
Hum Mutat,
27,
31-43.
|
 |
|
|
|
|
 |
L.Sun,
and
K.Warncke
(2006).
Comparative model of EutB from coenzyme B12-dependent ethanolamine ammonia-lyase reveals a beta8alpha8, TIM-barrel fold and radical catalytic site structural features.
|
| |
Proteins,
64,
308-319.
|
 |
|
|
|
|
 |
R.Banerjee,
A.Dybala-Defratyka,
and
P.Paneth
(2006).
Quantum catalysis in B12-dependent methylmalonyl-CoA mutase: experimental and computational insights.
|
| |
Philos Trans R Soc Lond B Biol Sci,
361,
1333-1339.
|
 |
|
|
|
|
 |
S.K.Moestrup
(2006).
New insights into carrier binding and epithelial uptake of the erythropoietic nutrients cobalamin and folate.
|
| |
Curr Opin Hematol,
13,
119-123.
|
 |
|
|
|
|
 |
A.Kambo,
V.S.Sharma,
D.E.Casteel,
V.L.Woods,
R.B.Pilz,
and
G.R.Boss
(2005).
Nitric oxide inhibits mammalian methylmalonyl-CoA mutase.
|
| |
J Biol Chem,
280,
10073-10082.
|
 |
|
|
|
|
 |
C.Acquaviva,
J.F.Benoist,
S.Pereira,
I.Callebaut,
T.Koskas,
D.Porquet,
and
J.Elion
(2005).
Molecular basis of methylmalonyl-CoA mutase apoenzyme defect in 40 European patients affected by mut(o) and mut- forms of methylmalonic acidemia: identification of 29 novel mutations in the MUT gene.
|
| |
Hum Mutat,
25,
167-176.
|
 |
|
|
|
|
 |
M.Fukuoka,
Y.Nakanishi,
R.B.Hannak,
B.Kräutler,
and
T.Toraya
(2005).
Homoadenosylcobalamins as probes for exploring the active sites of coenzyme B12-dependent diol dehydratase and ethanolamine ammonia-lyase.
|
| |
FEBS J,
272,
4787-4796.
|
 |
|
|
|
|
 |
R.J.Chandler,
and
C.P.Venditti
(2005).
Genetic and genomic systems to study methylmalonic acidemia.
|
| |
Mol Genet Metab,
86,
34-43.
|
 |
|
|
|
|
 |
F.Berkovitch,
E.Behshad,
K.H.Tang,
E.A.Enns,
P.A.Frey,
and
C.L.Drennan
(2004).
A locking mechanism preventing radical damage in the absence of substrate, as revealed by the x-ray structure of lysine 5,6-aminomutase.
|
| |
Proc Natl Acad Sci U S A,
101,
15870-15875.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
F.Berkovitch,
Y.Nicolet,
J.T.Wan,
J.T.Jarrett,
and
C.L.Drennan
(2004).
Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme.
|
| |
Science,
303,
76-79.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Korotkova,
and
M.E.Lidstrom
(2004).
MeaB is a component of the methylmalonyl-CoA mutase complex required for protection of the enzyme from inactivation.
|
| |
J Biol Chem,
279,
13652-13658.
|
 |
|
|
|
|
 |
D.M.Anstrom,
K.Kallio,
and
S.J.Remington
(2003).
Structure of the Escherichia coli malate synthase G:pyruvate:acetyl-coenzyme A abortive ternary complex at 1.95 A resolution.
|
| |
Protein Sci,
12,
1822-1832.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.K.Karpowich,
H.H.Huang,
P.C.Smith,
and
J.F.Hunt
(2003).
Crystal structures of the BtuF periplasmic-binding protein for vitamin B12 suggest a functionally important reduction in protein mobility upon ligand binding.
|
| |
J Biol Chem,
278,
8429-8434.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Shibata,
Y.Nakanishi,
M.Fukuoka,
M.Yamanishi,
N.Yasuoka,
and
T.Toraya
(2003).
Structural rationalization for the lack of stereospecificity in coenzyme B12-dependent diol dehydratase.
|
| |
J Biol Chem,
278,
22717-22725.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Banerjee,
and
S.W.Ragsdale
(2003).
The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes.
|
| |
Annu Rev Biochem,
72,
209-247.
|
 |
|
|
|
|
 |
B.Hao,
W.Gong,
T.K.Ferguson,
C.M.James,
J.A.Krzycki,
and
M.K.Chan
(2002).
A new UAG-encoded residue in the structure of a methanogen methyltransferase.
|
| |
Science,
296,
1462-1466.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Gruber,
and
C.Kratky
(2002).
Coenzyme B(12) dependent glutamate mutase.
|
| |
Curr Opin Chem Biol,
6,
598-603.
|
 |
|
|
|
|
 |
M.Cervantes,
and
F.J.Murillo
(2002).
Role for vitamin B(12) in light induction of gene expression in the bacterium Myxococcus xanthus.
|
| |
J Bacteriol,
184,
2215-2224.
|
 |
|
|
|
|
 |
M.D.Sintchak,
G.Arjara,
B.A.Kellogg,
J.Stubbe,
and
C.L.Drennan
(2002).
The crystal structure of class II ribonucleotide reductase reveals how an allosterically regulated monomer mimics a dimer.
|
| |
Nat Struct Biol,
9,
293-300.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Vlasie,
S.Chowdhury,
and
R.Banerjee
(2002).
Importance of the histidine ligand to coenzyme B12 in the reaction catalyzed by methylmalonyl-CoA mutase.
|
| |
J Biol Chem,
277,
18523-18527.
|
 |
|
|
|
|
 |
M.Yamanishi,
M.Yunoki,
T.Tobimatsu,
H.Sato,
J.Matsui,
A.Dokiya,
Y.Iuchi,
K.Oe,
K.Suto,
N.Shibata,
Y.Morimoto,
N.Yasuoka,
and
T.Toraya
(2002).
The crystal structure of coenzyme B12-dependent glycerol dehydratase in complex with cobalamin and propane-1,2-diol.
|
| |
Eur J Biochem,
269,
4484-4494.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Toraya
(2002).
Enzymatic radical catalysis: coenzyme B12-dependent diol dehydratase.
|
| |
Chem Rec,
2,
352-366.
|
 |
|
|
|
|
 |
E.N.Marsh,
and
C.L.Drennan
(2001).
Adenosylcobalamin-dependent isomerases: new insights into structure and mechanism.
|
| |
Curr Opin Chem Biol,
5,
499-505.
|
 |
|
|
|
|
 |
G.Garau,
S.N.Fedosov,
T.E.Petersen,
S.Geremia,
and
L.Randaccio
(2001).
Crystallization and preliminary X-ray diffraction analysis of human transcobalamin, a vitamin B12-transporting protein.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
1890-1892.
|
 |
|
|
|
|
 |
K.Gruber,
R.Reitzer,
and
C.Kratky
(2001).
Radical Shuttling in a Protein: Ribose Pseudorotation Controls Alkyl-Radical Transfer in the Coenzyme B(12) Dependent Enzyme Glutamate Mutase This work was supported by the Österreichische Akademie der Wissenschaften (APART fellowship 614), the Österreichische Fonds zur Förderung der wissenschaftlichen Forschung (FWF-project 11599), and the European Commission (TMR project number ERB 4061 PL 95-0307). Crystallographic data were collected at the EMBL-beamline BW7B at DESY in Hamburg, Germany. We thank the beamline scientists for their assistance, and Ingrid Dreveny, Günter Gartler, Gerwald Jogl, and Oliver Sauer for their help during data collection. This research emerged from a collaboration with Prof. W. Buckel (Marburg) who supplied us with clones of the glutamate mutase proteins.
|
| |
Angew Chem Int Ed Engl,
40,
3377-3380.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.A.Frey
(2001).
Radical mechanisms of enzymatic catalysis.
|
| |
Annu Rev Biochem,
70,
121-148.
|
 |
|
|
|
|
 |
P.M.Kozlowski
(2001).
Quantum chemical modeling of Co--C bond activation in B(12)-dependent enzymes.
|
| |
Curr Opin Chem Biol,
5,
736-743.
|
 |
|
|
|
|
 |
S.D.Wetmore,
D.M.Smith,
and
L.Radom
(2001).
Catalysis by mutants of methylmalonyl-CoA mutase: a theoretical rationalization for a change in the rate-determining step.
|
| |
Chembiochem,
2,
919-922.
|
 |
|
|
|
|
 |
W.Zhang,
and
K.A.Reynolds
(2001).
MeaA, a putative coenzyme B12-dependent mutase, provides methylmalonyl coenzyme A for monensin biosynthesis in Streptomyces cinnamonensis.
|
| |
J Bacteriol,
183,
2071-2080.
|
 |
|
|
|
|
 |
B.Hoffmann,
M.Oberhuber,
E.Stupperich,
H.Bothe,
W.Buckel,
R.Konrat,
and
B.Kräutler
(2000).
Native corrinoids from Clostridium cochlearium are adeninylcobamides: spectroscopic analysis and identification of pseudovitamin B(12) and factor A.
|
| |
J Bacteriol,
182,
4773-4782.
|
 |
|
|
|
|
 |
C.H.Chang,
and
P.A.Frey
(2000).
Cloning, sequencing, heterologous expression, purification, and characterization of adenosylcobalamin-dependent D-lysine 5, 6-aminomutase from Clostridium sticklandii.
|
| |
J Biol Chem,
275,
106-114.
|
 |
|
|
|
|
 |
B.Hoffmann,
R.Konrat,
H.Bothe,
W.Buckel,
and
B.Kräutler
(1999).
Structure and dynamics of the B12-binding subunit of glutamate mutase from Clostridium cochlearium.
|
| |
Eur J Biochem,
263,
178-188.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Masuda,
T.Yamaguchi,
T.Tobimatsu,
T.Toraya,
K.Suto,
N.Shibata,
Y.Morimoto,
Y.Higuchi,
and
N.Yasuoka
(1999).
Crystallization and preliminary x-ray study of two crystal forms of Klebsiella oxytoca diol dehydratase-cyanocobalamin complex.
|
| |
Acta Crystallogr D Biol Crystallogr,
55,
907-909.
|
 |
|
|
|
|
 |
N.Maiti,
L.Widjaja,
and
R.Banerjee
(1999).
Proton transfer from histidine 244 may facilitate the 1,2 rearrangement reaction in coenzyme B(12)-dependent methylmalonyl-CoA mutase.
|
| |
J Biol Chem,
274,
32733-32737.
|
 |
|
|
|
|
 |
N.Nagano,
E.G.Hutchinson,
and
J.M.Thornton
(1999).
Barrel structures in proteins: automatic identification and classification including a sequence analysis of TIM barrels.
|
| |
Protein Sci,
8,
2072-2084.
|
 |
|
|
|
|
 |
P.Langan,
M.Lehmann,
C.Wilkinson,
G.Jogl,
and
C.Kratky
(1999).
Neutron Laue diffraction studies of coenzyme cob(II)alamin.
|
| |
Acta Crystallogr D Biol Crystallogr,
55,
51-59.
|
 |
|
|
|
|
 |
T.Izard,
and
A.Geerlof
(1999).
The crystal structure of a novel bacterial adenylyltransferase reveals half of sites reactivity.
|
| |
EMBO J,
18,
2021-2030.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.C.Lawrence,
and
J.Stubbe
(1998).
The function of adenosylcobalamin in the mechanism of ribonucleoside triphosphate reductase from Lactobacillus leichmannii.
|
| |
Curr Opin Chem Biol,
2,
650-655.
|
 |
|
|
|
|
 |
C.E.Adjalla,
A.R.Hosack,
B.M.Gilfix,
E.Lamothe,
S.Sun,
A.Chan,
S.Evans,
N.V.Matiaszuk,
and
D.S.Rosenblatt
(1998).
Seven novel mutations in mut methylmalonic aciduria.
|
| |
Hum Mutat,
11,
270-274.
|
 |
|
|
|
|
 |
K.Yamada,
T.Tobimatsu,
and
T.Toraya
(1998).
Cloning, sequencing, and heterologous expression of rat methionine synthase cDNA.
|
| |
Biosci Biotechnol Biochem,
62,
2155-2160.
|
 |
|
|
|
|
 |
K.Zerbe-Burkhardt,
A.Ratnatilleke,
N.Philippon,
A.Birch,
A.Leiser,
J.W.Vrijbloed,
D.Hess,
P.Hunziker,
and
J.A.Robinson
(1998).
Cloning, sequencing, expression, and insertional inactivation of the gene for the large subunit of the coenzyme B12-dependent isobutyryl-CoA mutase from Streptomyces cinnamonensis.
|
| |
J Biol Chem,
273,
6508-6517.
|
 |
|
|
|
|
 |
P.K.Mishra,
and
D.G.Drueckhammer
(1998).
Novel cofactor derivatives and cofactor-based models.
|
| |
Curr Opin Chem Biol,
2,
758-765.
|
 |
|
|
|
|
 |
R.Daniel,
T.A.Bobik,
and
G.Gottschalk
(1998).
Biochemistry of coenzyme B12-dependent glycerol and diol dehydratases and organization of the encoding genes.
|
| |
FEMS Microbiol Rev,
22,
553-566.
|
 |
|
|
|
|
 |
A.Abend,
V.Illich,
and
J.Rétey
(1997).
Further insights into the mechanism of action of methylmalonyl-CoA mutase by electron paramagnetic resonance studies.
|
| |
Eur J Biochem,
249,
180-186.
|
 |
|
|
|
|
 |
L.Poppe,
E.Stupperich,
W.E.Hull,
T.Buckel,
and
J.Rétey
(1997).
A base-off analogue of coenzyme-B12 with a modified nucleotide loop--1H-NMR structure analysis and kinetic studies with (R)-methylmalonyl-CoA mutase, glycerol dehydratase, and diol dehydratase.
|
| |
Eur J Biochem,
250,
303-307.
|
 |
|
|
|
|
 |
L.Poppe,
and
J.Rétey
(1997).
Kinetic investigations with inhibitors that mimic the posthomolysis intermediate in the reactions of coenzyme-B12-dependent glycerol dehydratase and diol dehydratase.
|
| |
Eur J Biochem,
245,
398-401.
|
 |
|
|
|
|
 |
M.L.Ludwig,
and
R.G.Matthews
(1997).
Structure-based perspectives on B12-dependent enzymes.
|
| |
Annu Rev Biochem,
66,
269-313.
|
 |
|
|
|
|
 |
P.A.Frey
(1997).
Radicals in enzymatic reactions.
|
| |
Curr Opin Chem Biol,
1,
347-356.
|
 |
|
|
|
|
 |
P.T.Erskine,
N.Senior,
S.Awan,
R.Lambert,
G.Lewis,
I.J.Tickle,
M.Sarwar,
P.Spencer,
P.Thomas,
M.J.Warren,
P.M.Shoolingin-Jordan,
S.P.Wood,
and
J.B.Cooper
(1997).
X-ray structure of 5-aminolaevulinate dehydratase, a hybrid aldolase.
|
| |
Nat Struct Biol,
4,
1025-1031.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Banerjee
(1997).
The Yin-Yang of cobalamin biochemistry.
|
| |
Chem Biol,
4,
175-186.
|
 |
|
|
|
|
 |
U.Ermler,
W.Grabarse,
S.Shima,
M.Goubeaud,
and
R.K.Thauer
(1997).
Crystal structure of methyl-coenzyme M reductase: the key enzyme of biological methane formation.
|
| |
Science,
278,
1457-1462.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Volbeda,
J.C.Fontecilla-Camps,
and
M.Frey
(1996).
Novel metal sites in protein structures.
|
| |
Curr Opin Struct Biol,
6,
804-812.
|
 |
|
|
|
|
 |
C.Engel,
and
R.Wierenga
(1996).
The diverse world of coenzyme A binding proteins.
|
| |
Curr Opin Struct Biol,
6,
790-797.
|
 |
|
|
|
|
 |
C.K.Engel,
M.Mathieu,
J.P.Zeelen,
J.K.Hiltunen,
and
R.K.Wierenga
(1996).
Crystal structure of enoyl-coenzyme A (CoA) hydratase at 2.5 angstroms resolution: a spiral fold defines the CoA-binding pocket.
|
| |
EMBO J,
15,
5135-5145.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.L.Drennan,
R.G.Matthews,
D.S.Rosenblatt,
F.D.Ledley,
W.A.Fenton,
and
M.L.Ludwig
(1996).
Molecular basis for dysfunction of some mutant forms of methylmalonyl-CoA mutase: deductions from the structure of methionine synthase.
|
| |
Proc Natl Acad Sci U S A,
93,
5550-5555.
|
 |
|
|
|
|
 |
D.E.Holloway,
H.P.Chen,
and
E.N.Marsh
(1996).
Carboxymethylation of MutS-cysteine-15 specifically inactivates adenosylcobalamin-dependent glutamate mutase. Examination of the role of this residue in coenzyme-binding and catalysis.
|
| |
J Biol Chem,
271,
29121-29125.
|
 |
|
|
|
|
 |
N.H.Thomä,
and
P.F.Leadlay
(1996).
Homology modeling of human methylmalonyl-CoA mutase: a structural basis for point mutations causing methylmalonic aciduria.
|
| |
Protein Sci,
5,
1922-1927.
|
 |
|
|
|
|
 |
T.Tobimatsu,
M.Azuma,
H.Matsubara,
H.Takatori,
T.Niida,
K.Nishimoto,
H.Satoh,
R.Hayashi,
and
T.Toraya
(1996).
Cloning, sequencing, and high level expression of the genes encoding adenosylcobalamin-dependent glycerol dehydrase of Klebsiella pneumoniae.
|
| |
J Biol Chem,
271,
22352-22357.
|
 |
|
|
|
|
 |
U.Harms,
and
R.K.Thauer
(1996).
The corrinoid-containing 23-kDa subunit MtrA of the energy-conserving N5-methyltetrahydromethanopterin:coenzyme M methyltransferase complex from Methanobacterium thermoautotrophicum. EPR spectroscopic evidence for a histidine residue as a cobalt ligand of the cobamide.
|
| |
Eur J Biochem,
241,
149-154.
|
 |
|
|
|
|
 |
W.G.Hol
(1996).
The sophisticated masters of the cell.
|
| |
Curr Opin Struct Biol,
6,
777-780.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

