Homogentisate 1,2-dioxygenase
Homogentistate dioxygenase (HGO) catalyses the metabolic degredation of Phe and Tyr amino acids. The ring opening reaction requires non-heme Fe(2+) to incorporate both atoms of molecular oxygen into homogentisate. The accumulation of this substrate, when insufficient levels of HGO are present, results in the deposition of of insoluble ochromotic pigments in conective tissues, leading to degenerative arthritis.
Reference Protein and Structure
- Sequence
-
Q93099
 (1.13.11.5)
(1.13.11.5)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
1ey2
- HUMAN HOMOGENTISATE DIOXYGENASE WITH FE(II)
(2.3 Å)



- Catalytic CATH Domains
-
2.60.120.10
 (see all for 1ey2)
(see all for 1ey2)
- Cofactors
- Iron(2+) (1)
Enzyme Reaction (EC:1.13.11.5)
Enzyme Mechanism
Introduction
Direct bidentate coordination occurs between Fe(2+) and the homogentisate 1-acetate and 2-hydroxylate groups, while His 292 hydrogen bonds to to the 5-hydroxyl group. The essential metal ion is coordinated to His 371, His 335 and Glu 341. Molecular dioxygen binds to iron and reacts with homogentisate, forming a peroxo-bridged intermediate. The second step is a homolytic cleave of the O-O bond in this intermediate, which generates an arene oxide radical. Lastly, nucleophilic attack of the iron bound OH group at the carbonyl of the arene oxide radical, opening the epoxide ring. In the mechanism proposed above, His 292 is expected to remain neutral, limiting its role to substrate binding. However, a second mechanism, higher in energy, proposed by Borowski et al. implicates His 292 as a general acid/base in a proton assisted Criegee Rearrangement for HGO.
Catalytic Residues Roles
| UniProt | PDB* (1ey2) | ||
| His292 | His292(318)A | Helps activate and stabilise the reactive intermediates and transition states formed during the course of the reaction. | enhance reactivity, electrostatic stabiliser |
| His335, Glu341, His371 | His335(361)A, Glu341(367)A, His371(397)A | Forms part of the catalytic iron binding site. | metal ligand |
| His365 | His365(391)A | Acts as a general acid/base during the course of the reaction. | proton acceptor, proton donor |
Chemical Components
bimolecular nucleophilic addition, overall reactant used, proton transfer, electron transfer, homolysis, intramolecular homolytic addition, intramolecular homolytic elimination, native state of cofactor regenerated, overall product formed, inferred reaction step, native state of enzyme regeneratedReferences
- Borowski T et al. (2005), J Am Chem Soc, 127, 17303-17314. Catalytic Reaction Mechanism of Homogentisate Dioxygenase: A Hybrid DFT Study. DOI:10.1021/ja054433j. PMID:16332080.
- Buongiorno D et al. (2013), Coord Chem Rev, 257, 541-563. Structure and function of atypically coordinated enzymatic mononuclear non-heme-Fe(II) centers. DOI:10.1016/j.ccr.2012.04.028. PMID:24850951.
- Titus GP et al. (2000), Nat Struct Biol, 7, 542-546. Crystal structure of human homogentisate dioxygenase. DOI:10.1038/76756. PMID:10876237.
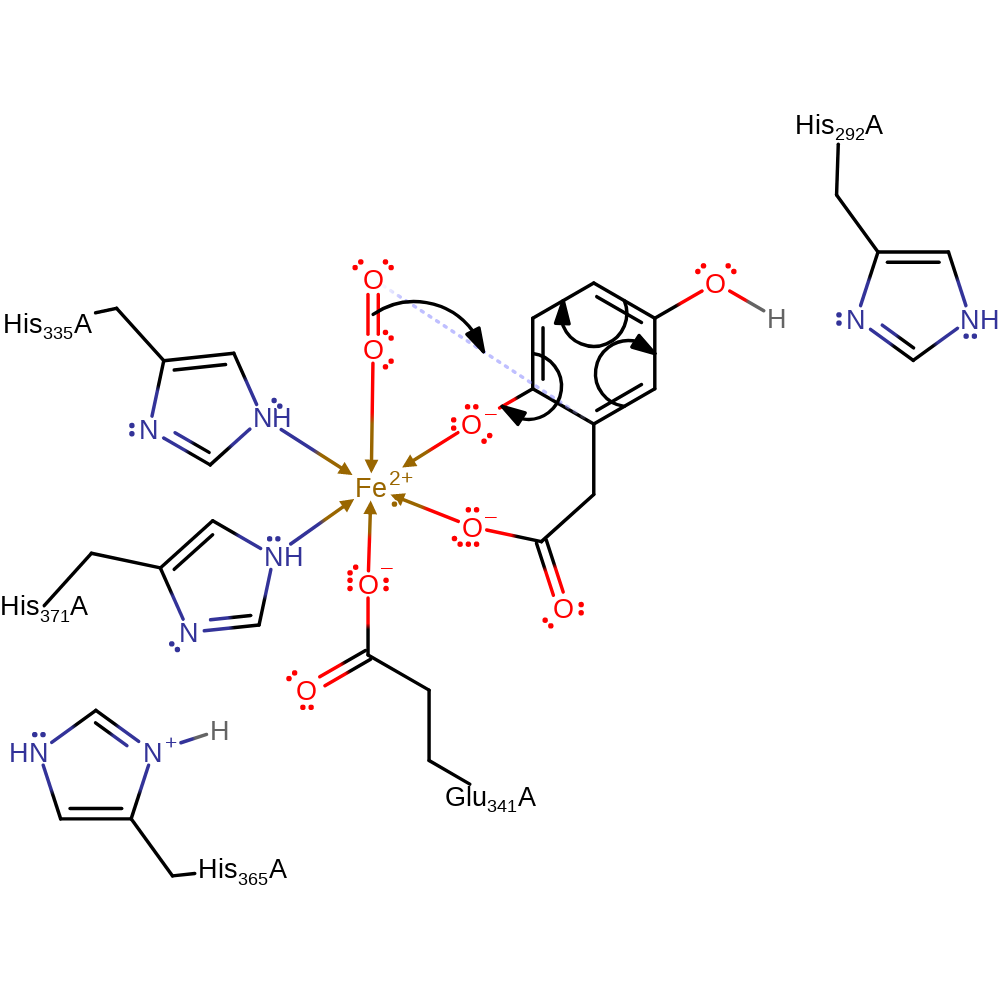
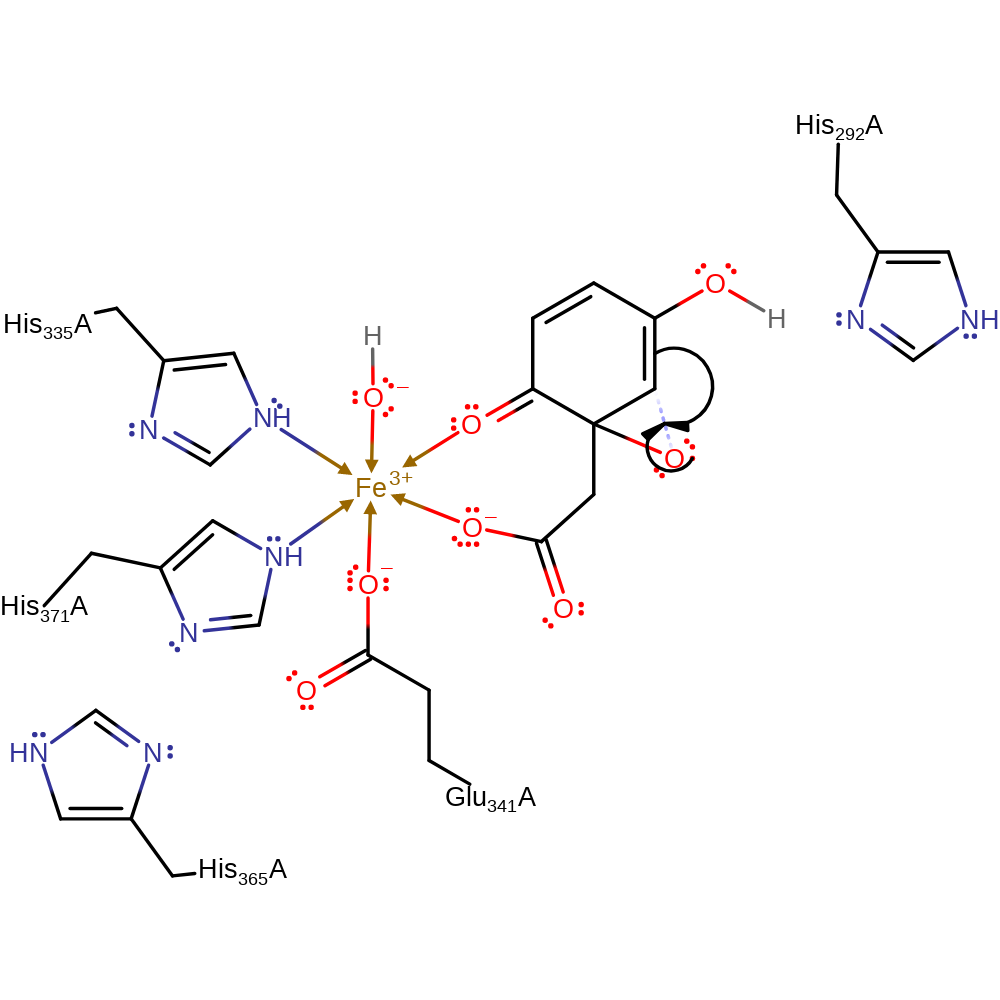
Step 1. The iron-bound dioxygen attacks the aromatic ring at the C2 carbon
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu341(367)A | metal ligand |
| His371(397)A | metal ligand |
| His335(361)A | metal ligand |
| His292(318)A | enhance reactivity |
Chemical Components
ingold: bimolecular nucleophilic addition, overall reactant usedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu341(367)A | metal ligand |
| His371(397)A | metal ligand |
| His335(361)A | metal ligand |
| His292(318)A | enhance reactivity |
| His365(391)A | proton donor |
Chemical Components
proton transfer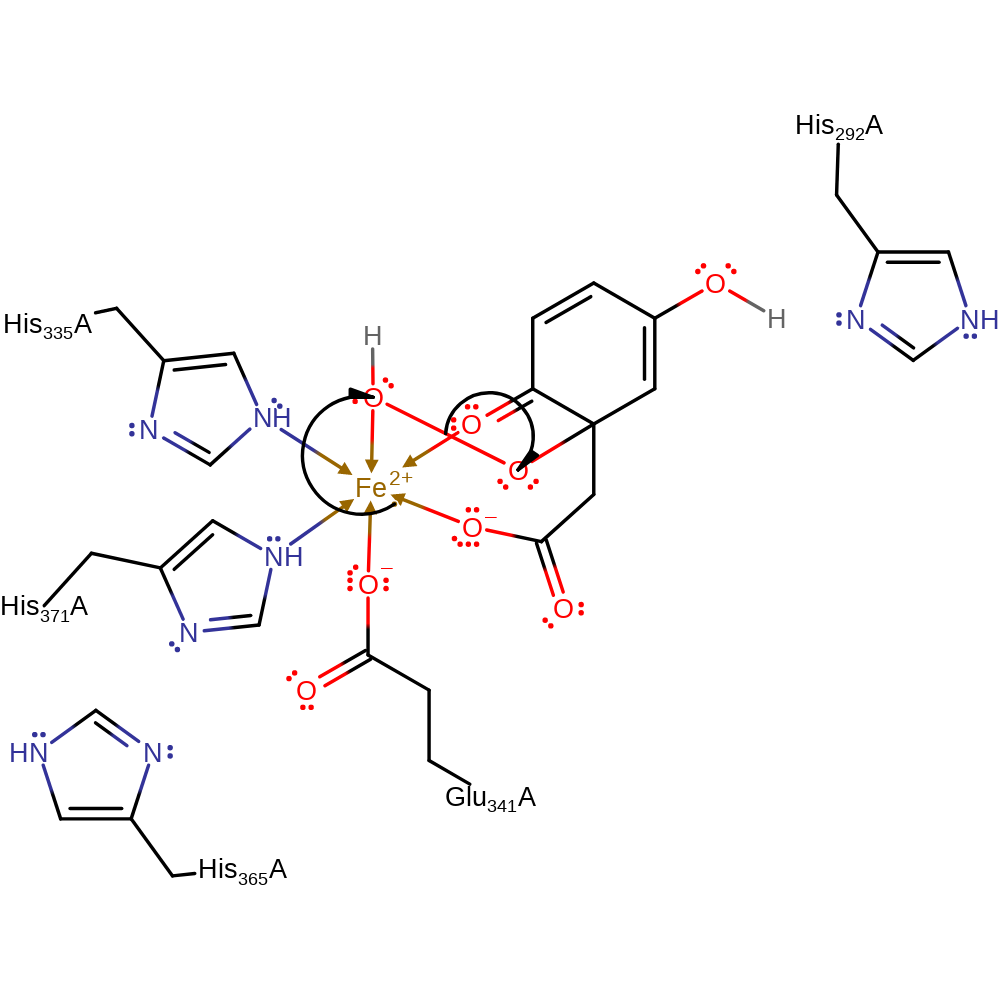
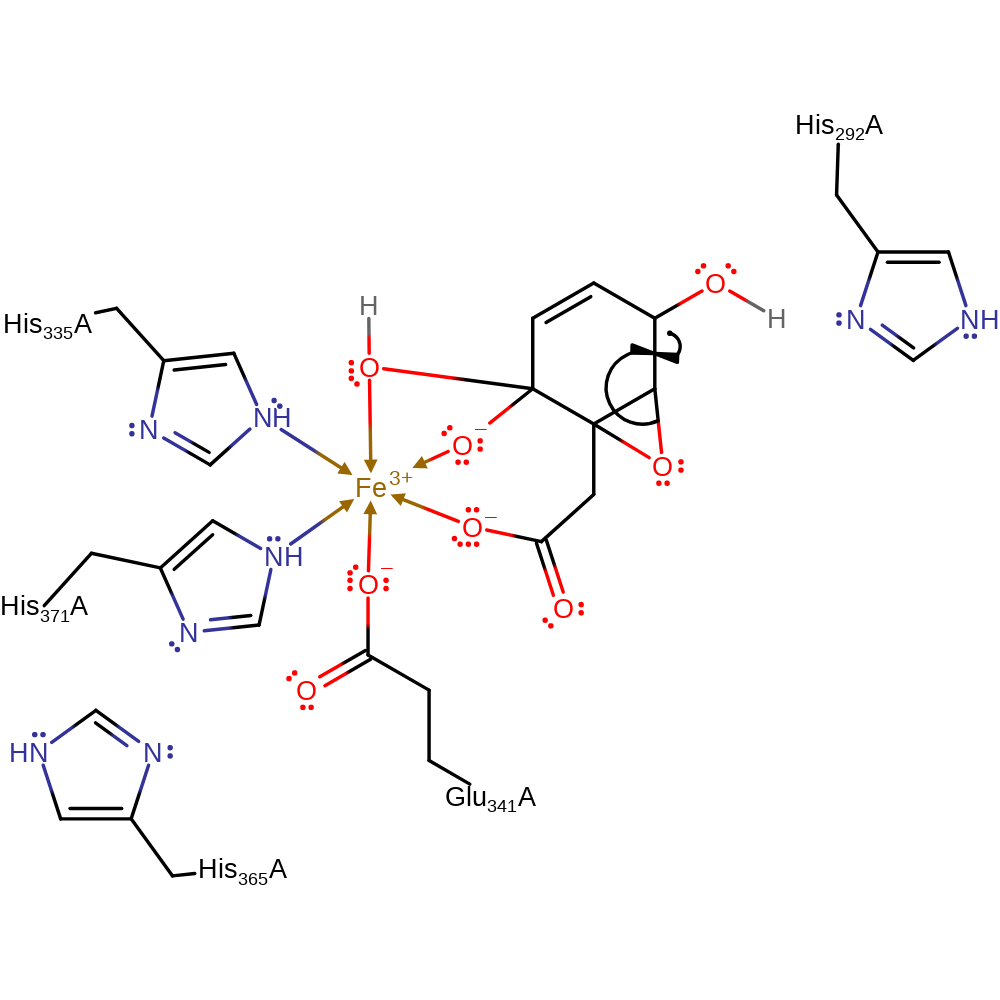
Step 3. A single electron from the iron(II) ion is donated to the peroxo group, which initiates the homolytic cleavage of the peroxo bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu341(367)A | metal ligand |
| His371(397)A | metal ligand |
| His335(361)A | metal ligand |
| His292(318)A | enhance reactivity |
Chemical Components
electron transfer, homolysisCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His292(318)A | enhance reactivity |
| Glu341(367)A | metal ligand |
| His371(397)A | metal ligand |
| His335(361)A | metal ligand |
Chemical Components
ingold: intramolecular homolytic addition
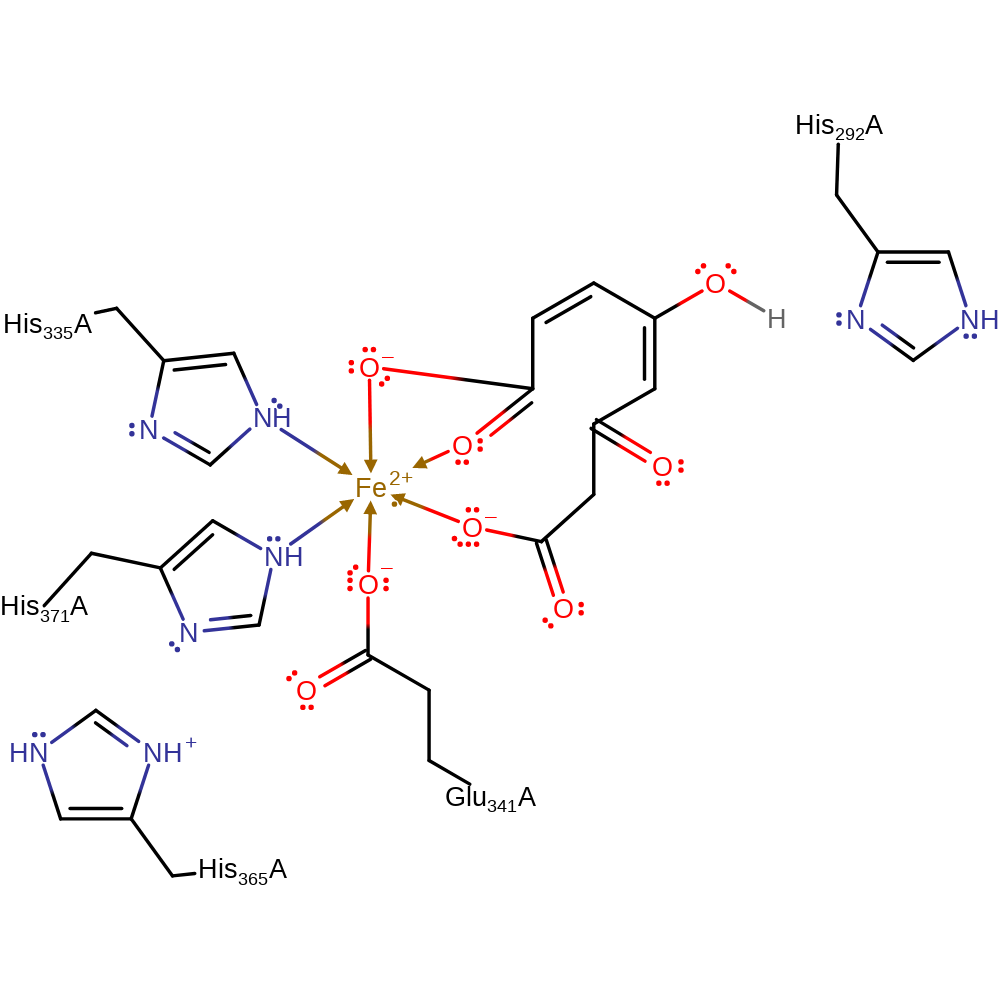
Step 5. The hydroxide group attacks the carbonyl group bound to the iron(III) ion to form the new carboxylic acid group.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu341(367)A | metal ligand |
| His371(397)A | metal ligand |
| His335(361)A | metal ligand |
| His292(318)A | enhance reactivity |
Chemical Components
ingold: bimolecular nucleophilic additionCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu341(367)A | metal ligand |
| His371(397)A | metal ligand |
| His335(361)A | metal ligand |
| His292(318)A | enhance reactivity |
Chemical Components
ingold: intramolecular homolytic elimination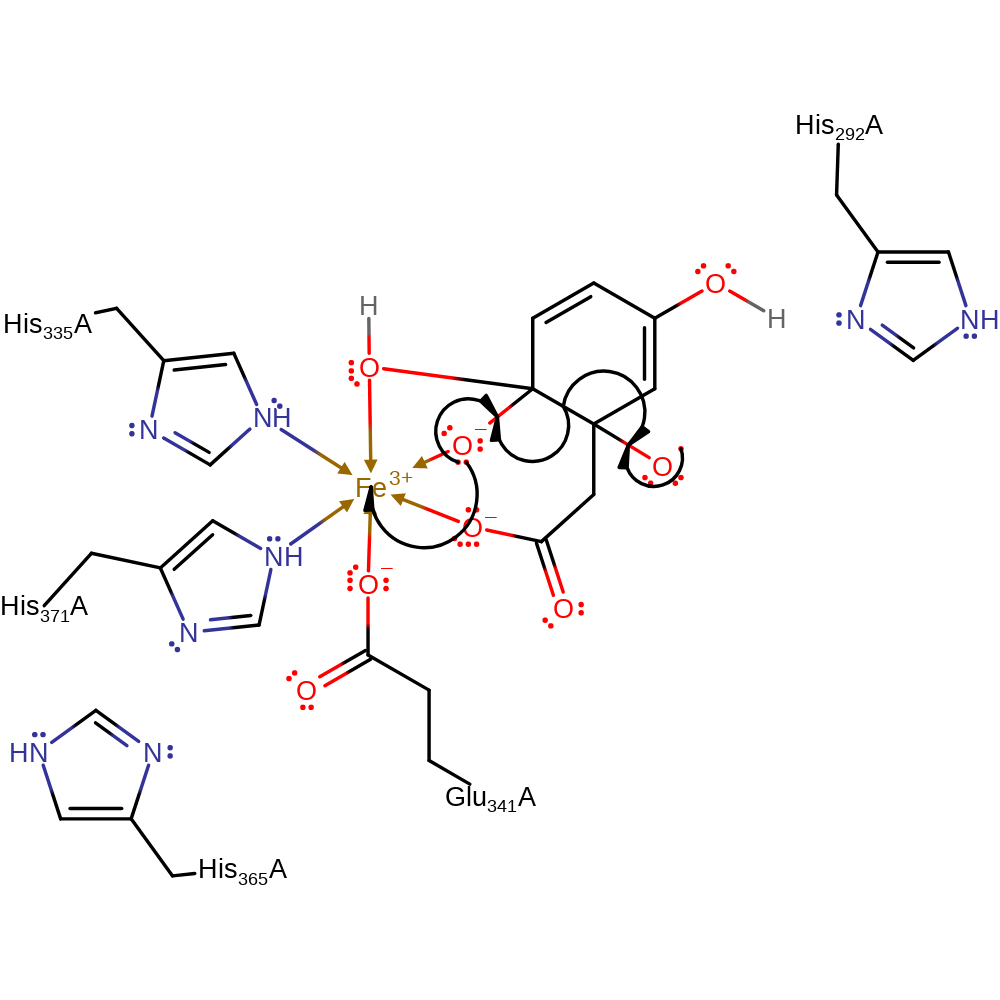
Step 7. The hydroxyl radical initiates a homolytic rearrangement that results in the formation of the final product and a single electron transfer to the Fe(III) centre to regenerate the cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu341(367)A | metal ligand |
| His371(397)A | metal ligand |
| His335(361)A | metal ligand |
| His292(318)A | enhance reactivity |
Chemical Components
ingold: intramolecular homolytic elimination, native state of cofactor regenerated, overall product formed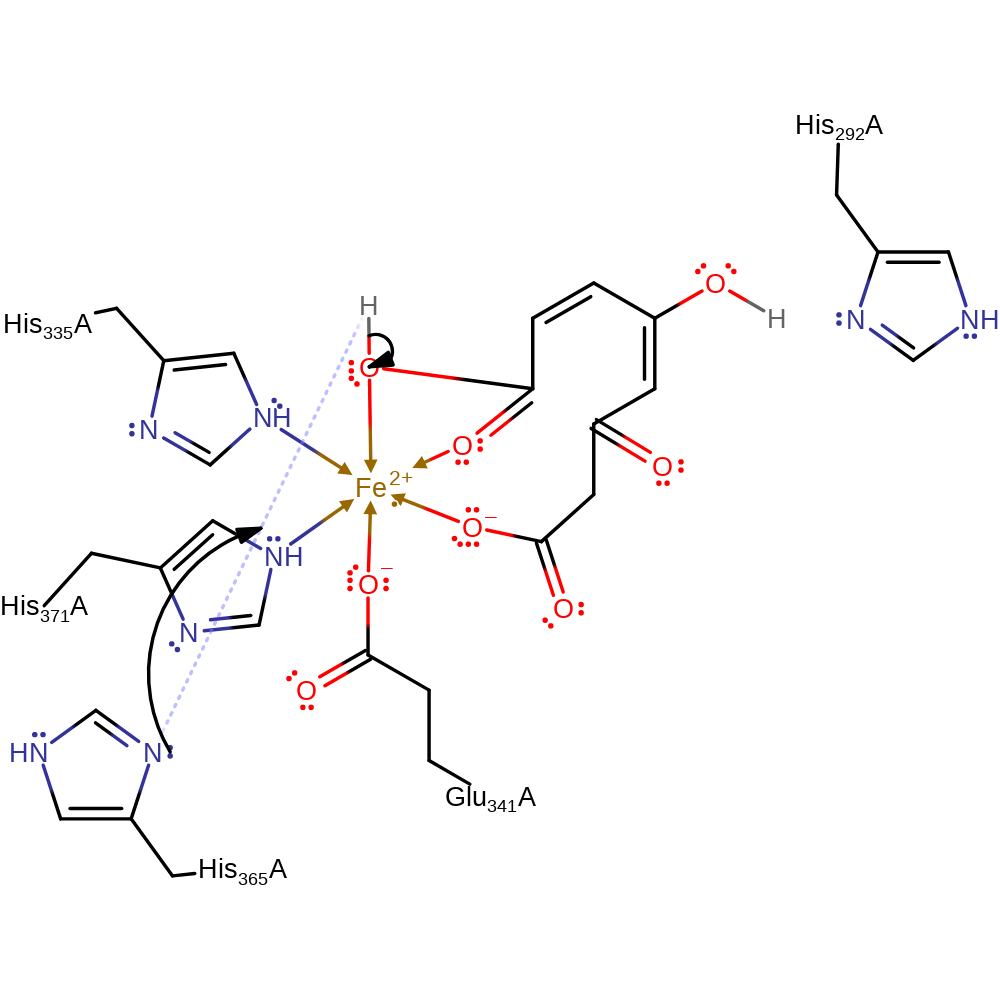
Step 8. Inferred step in which His365 abstracts a proton from the product to regenerate the active site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu341(367)A | metal ligand |
| His371(397)A | metal ligand |
| His335(361)A | metal ligand |
| His292(318)A | electrostatic stabiliser |
| His365(391)A | proton acceptor |





 Download:
Download: