DNA topoisomerase I (type 1A)
DNA topoisomerase enzymes modify DNA super-helicity and perform DNA decatenation using scission, manipulation and ligation reactions. Two families of topoisomerases exist, group I and II, cleaving single and double stranded DNA respectively. Each family can be subdivided into A and B groups, which represent subsets of proteins with no similarity in structure or sequence. All topoisomerases use a tyrosine to cleave DNA, forming a short-lived phosphotyrosyl intermediate. These bacterial enzymes reduce the topological stress in the DNA structure by relaxing negatively, but not positively, supercoiled DNA. Different classes of enzyme relax DNA to different extents and, for the 1A enzymes, activity correlates with the degree of DNA substrate superhelicity.
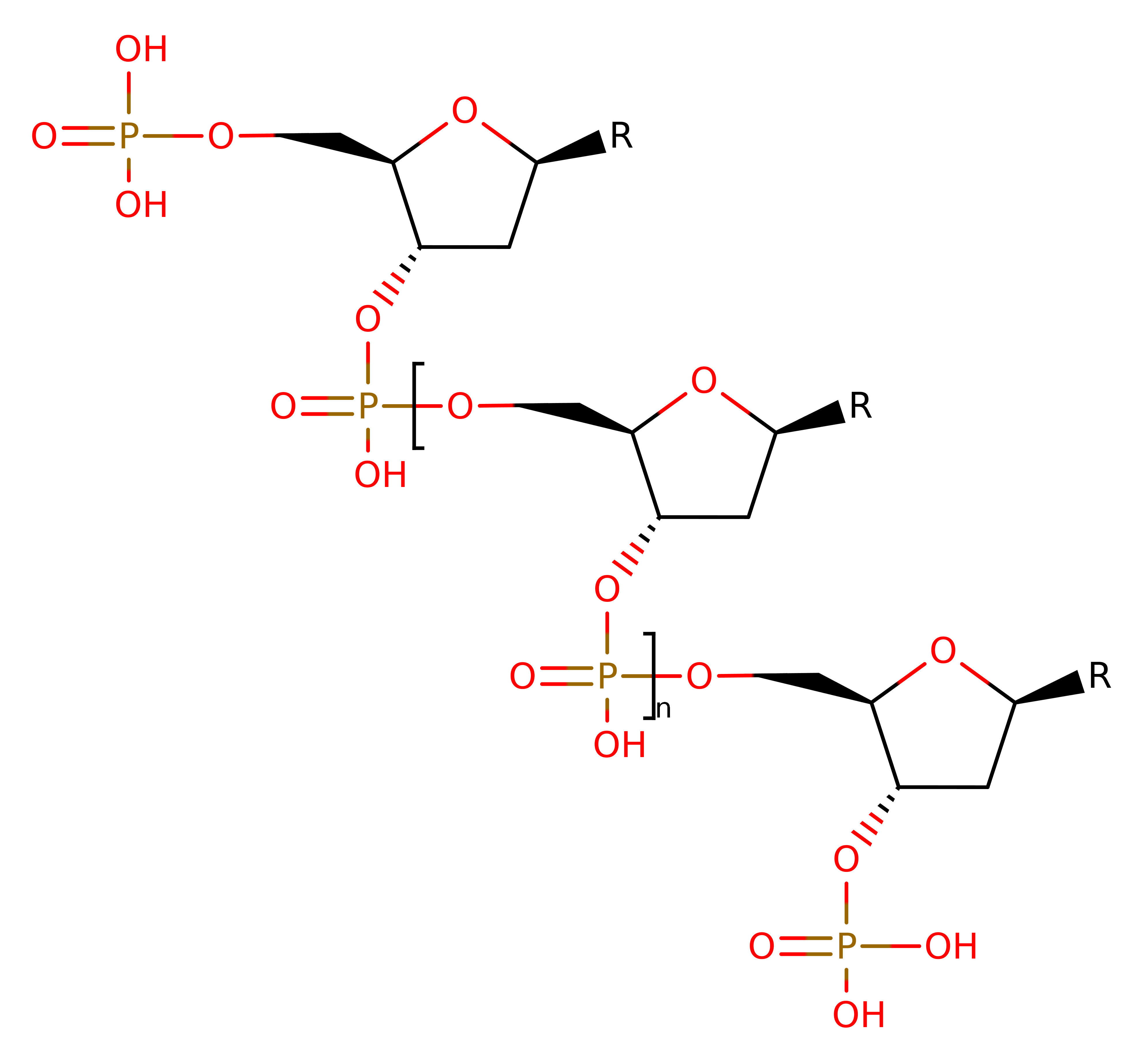
Type I DNA topoisomerase enzymes alter the topology of DNA by transiently breaking one or two strands of DNA, passing a single or double strand through the break and then resealing the break. This allows for the interconversion of topological isomers, which is necessary for a number of cellular transactions such as replication, transcription and recombination. The enzyme is a member of the IA sub family, classified by the covalently bound enzyme-DNA intermediate.
Unlike group II enzymes, the reactions catalysed by Escherichia coli topoisomerase are not coupled to binding or hydrolysis of ATP. Full-length topoisomerase I from Escherichia coli is a 97kDa metalloprotein containing three Zn(II) cations at the C-terminus. The structure annotated is a 67kDa, N-terminal truncation protein. While the tetra-cysteine motifs, thought to bind Zn(II) are not required for single strand DNA cleavage, without them the protein cannot relax negatively supercoiled DNA. However, the substrate specificity and reaction characteristics of the N-terminal truncation fragment are thought to parallel the full length and Zn depleted enzyme.
Reference Protein and Structure
- Sequence
-
P06612
 (5.6.2.1)
(5.6.2.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1ecl
- AMINO TERMINAL 67KDA DOMAIN OF ESCHERICHIA COLI DNA TOPOISOMERASE I (RESIDUES 2-590 OF MATURE PROTEIN) CLONING ARTIFACT ADDS TWO RESIDUES TO THE AMINO-TERMINUS WHICH WERE NOT OBSERVED IN THE EXPERIMENTAL ELECTRON DENSITY (GLY-2, SER-1).
(1.9 Å)



- Catalytic CATH Domains
-
1.10.290.10
 3.40.50.140
3.40.50.140  (see all for 1ecl)
(see all for 1ecl)
- Cofactors
- Magnesium(2+) (1)
Enzyme Reaction (EC:5.6.2.1)
Enzyme Mechanism
Introduction
The enzyme mechanism is generally accepted to proceed via two sequential trans-esterification reactions. Initially, the enzyme recognises and binds a DNA region, positioning the DNA in the active site. Cleavage of the single stranded DNA occurs via the nucleophilic attack of the Tyr 319 hydroxyl group at the 5' phosphoryl end, with the formation of a covalent bond between the two, while the 3' end remains non-covalently bound to the enzyme. The enzyme opens to allow the passage of the other strand through the gap created by separating the broken ends of the DNA. The gap closes, trapping the DNA strand, and acts to re-ligate the cleaved strand. The enzyme then opens to release both the re-ligated strand and the strand held in the transient gap.
Catalytic Residues Roles
| UniProt | PDB* (1ecl) | ||
| Arg321 | Arg321A | The positively charged side chain interacts with the oxygen of scissile phosphate group, and is implicated in the modification of the Tyr 319 pKa by stabilising the charged phenolate intermediate. | electrostatic stabiliser |
| Asp113, Glu115 | Asp113A, Glu115A | Coordinate the Mg ion | metal ligand |
| Tyr319 | Tyr319A | The OH group of the residue acts as a nucleophile towards an internucleoside phosphate group, resulting in a covalently bound enzyme-DNA intermediate. It has been suggested that the residue is deprotonated before nucleophilic attack occurs, although no general base has been identified. The proton is transferred to the deoxyribose 3' oxygen of the DNA substrate from the scissile phosphate. The residue interacts with the positively charged side chain of Arg 321, modifying its associated pKa and stabilising the charged phenolate intermediate. | covalently attached, hydrogen bond acceptor, nucleofuge, nucleophile, proton acceptor, proton donor, activator |
| Asp111, His365, Glu9 | Asp111A, His365A, Glu9A | The residue acts as a general base in proton removal from the 3' hydroxyl group during DNA rejoining, and a proton donor during DNA breakage. The large distance between the residue and the catalytic Tyr 319 suggest that it does not act as a direct base to the nucleophile. This residue is activate to act as a general acid/base by a proton relay through Asp111 and His365 (with His365 as the initiator). | hydrogen bond acceptor, hydrogen bond donor, metal ligand, proton acceptor, proton donor, proton relay, electrostatic stabiliser, increase acidity |
Chemical Components
proton transfer, overall reactant used, intermediate formation, enzyme-substrate complex formation, bimolecular nucleophilic substitution, intermediate collapse, overall product formed, native state of enzyme regenerated, inferred reaction stepReferences
- Bugreev DV et al. (2009), Biochemistry (Mosc), 74, 1467-1481. Structure and mechanism of action of type IA DNA topoisomerases. DOI:10.1134/s0006297909130045. PMID:20210704.
- Sissi C et al. (2009), Nucleic Acids Res, 37, 702-711. Effects of magnesium and related divalent metal ions in topoisomerase structure and function. DOI:10.1093/nar/gkp024. PMID:19188255.
- Perry K et al. (2002), J Biol Chem, 277, 13237-13245. Biochemical Characterization of an Invariant Histidine Involved in Escherichia coli DNA Topoisomerase I Catalysis. DOI:10.1074/jbc.m112019200. PMID:11809772.
- Chen SJ et al. (1998), J Biol Chem, 273, 6050-6056. Identification of Active Site Residues in Escherichia coli DNA Topoisomerase I. DOI:10.1074/jbc.273.11.6050. PMID:9497321.
- Lima CD et al. (1994), Nature, 367, 138-146. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. DOI:10.1038/367138a0. PMID:8114910.

Step 1. Cleavage of the single-stranded DNA via the formation of a covalent bond between then 5’-phosphoryl group and the hydroxyl of the active site tyrosine. The 3’ end of the DNA remains non-covalently bound to the enzyme.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu9A | electrostatic stabiliser, hydrogen bond donor, increase acidity |
| Asp111A | electrostatic stabiliser, hydrogen bond donor, increase acidity |
| His365A | electrostatic stabiliser, hydrogen bond acceptor, increase acidity |
| Arg321A | electrostatic stabiliser |
| Glu9A | metal ligand |
| Asp111A | metal ligand |
| Asp113A | metal ligand |
| Glu115A | metal ligand |
| Tyr319A | proton donor, nucleophile |
Chemical Components
proton transfer, overall reactant used, intermediate formation, enzyme-substrate complex formation, ingold: bimolecular nucleophilic substitution
Step 2. Cleavage of the single-stranded DNA via the formation of a covalent bond between then 5'-phosphoryl group and the hydroxyl of the active site tyrosine. The 3' end of the DNA remains non-covalently bound to the enzyme.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu9A | electrostatic stabiliser, hydrogen bond donor, increase acidity |
| Asp111A | electrostatic stabiliser, hydrogen bond donor, increase acidity |
| Tyr319A | activator, covalently attached |
| His365A | electrostatic stabiliser, hydrogen bond acceptor, increase acidity |
| Arg321A | electrostatic stabiliser |
| Glu9A | metal ligand |
| Asp111A | metal ligand |
| Asp113A | metal ligand |
| Glu115A | metal ligand |
| Glu9A | proton acceptor |
| His365A | proton acceptor |
| Asp111A | proton acceptor, proton donor |
| Glu9A | proton donor |
| Tyr319A | nucleofuge |
| Glu9A | proton relay |
| Asp111A | proton relay |
Chemical Components
proton transfer, intermediate collapse, enzyme-substrate complex formation, overall product formed, ingold: bimolecular nucleophilic substitution
Step 3. Tyr319 is reprotonated by His375, through a relay of acidic residues within the active site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu9A | hydrogen bond acceptor, hydrogen bond donor |
| Asp111A | hydrogen bond acceptor, hydrogen bond donor |
| Tyr319A | activator, hydrogen bond acceptor |
| His365A | hydrogen bond donor |
| Glu9A | metal ligand |
| Asp111A | metal ligand |
| Asp113A | metal ligand |
| Glu115A | metal ligand |
| Glu9A | proton donor, proton acceptor |
| Asp111A | proton acceptor |
| His365A | proton donor |
| Tyr319A | proton acceptor |
| Asp111A | proton donor |
| Glu9A | proton relay |
| Asp111A | proton relay |
 Download:
Download: