Indole-3-glycerol-phosphate synthase
Indole-3-glycerol phosphate synthase (EC:4.1.1.48) (IGPS) catalyses the fourth step in the biosynthesis of tryptophan, the ring closure of 1-(2-carboxy-phenylamino)-1-deoxyribulose into indol-3-glycerol-phosphate. In some bacteria, IGPS is a single chain enzyme. In others, such as Escherichia coli, it is the N-terminal domain of a bifunctional enzyme that also catalyses N-(5'-phosphoribosyl)anthranilate isomerase (EC:5.3.1.24) (PRAI) activity. In fungi, IGPS is the central domain of a trifunctional enzyme that contains a PRAI C-terminal domain and a glutamine amidotransferase (EC:2.4.2) (GATase) N-terminal domain.
Reference Protein and Structure
- Sequence
-
Q06121
 (4.1.1.48)
(4.1.1.48)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Sulfolobus solfataricus P2 (Archaea)

- PDB
-
1igs
- INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS AT 2.0 A RESOLUTION
(2.0 Å)



- Catalytic CATH Domains
-
3.20.20.70
 (see all for 1igs)
(see all for 1igs)
Enzyme Reaction (EC:4.1.1.48)
Enzyme Mechanism
Introduction
Indoleglycerol phosphate synthase catalyzes the ring closure of an N-alkylated anthranilate to a 3-alkyl indole derivative. There is still considerable debate as to the exact roles of the residues involved in this mechanism, however the basics remain the same:
- The lone pair of the nitrogen atom in the substrate initiate the ring closure reaction with concomitant deprotonation of Lys110
- Carbon dioxide is eliminated
- The dehydration reaction occurs, this is most likely initiated by Glu51 abstracting a proton from the nitrogen atom of the newly formed imidazole ring. Other suggestions have included Glu159 or Glu210 as the general base in this step. The eliminated hydroxyl group abstracts a proton from Lys53
- The enzyme is returned to its ground protonation state. This likely occurs through bulk solvent.
Catalytic Residues Roles
| UniProt | PDB* (1igs) | ||
| Glu51 | Glu51A | General base in dehydration step. | hydrogen bond acceptor, proton acceptor, proton donor, electrostatic stabiliser, increase acidity, increase basicity |
| Glu210 | Glu210A | There is some debate as to the exact role of this residue and Glu159. Computational studies had suggested that this was one the general base in the ring closure step, however work from 2013 suggested that this residue is involved in substrate binding and stabilisation. | electrostatic stabiliser |
| Asn180, Ser211 | Asn180A, Ser211A | Forms part of a Ser-Asn-Glu triad, responsible for activating the glutamate to act as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser, increase acidity |
| Lys53 | Lys53A | General acid in dehydration step. | proton acceptor, hydrogen bond donor, electrostatic stabiliser, proton donor |
| Lys110 | Lys110A | General acid during the ring closure reaction. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Glu159 | Glu159A | There is some debate as to the exact role of this residue and Glu210. Computational studies had suggested that E159 was the general base in the ring closure step, however work from 2013 has suggested that this residue is involved in stabilising and activating the intermediate for the ring closure step. | activator, increase nucleophilicity, hydrogen bond acceptor |
Chemical Components
intramolecular nucleophilic addition, proton transfer, cyclisation, unimolecular elimination by the conjugate base, decarboxylation, intramolecular elimination, dehydration, inferred reaction step, native state of enzyme regeneratedReferences
- Zaccardi MJ et al. (2013), J Biol Chem, 288, 26350-26356. Functional Identification of the General Acid and Base in the Dehydration Step of Indole-3-glycerol Phosphate Synthase Catalysis. DOI:10.1074/jbc.m113.487447. PMID:23900843.
- Zaccardi MJ et al. (2014), Protein Sci, 23, 302-311. Loop-loop interactions govern multiple steps in indole-3-glycerol phosphate synthase catalysis. DOI:10.1002/pro.2416. PMID:24403092.
- Schlee S et al. (2013), Biochemistry, 52, 132-142. Kinetic Mechanism of Indole-3-glycerol Phosphate Synthase. DOI:10.1021/bi301342j. PMID:23214473.
- Czekster CM et al. (2009), Arch Biochem Biophys, 486, 19-26. Steady-state kinetics of indole-3-glycerol phosphate synthase from Mycobacterium tuberculosis. DOI:10.1016/j.abb.2009.04.001. PMID:19364491.
- Mazumder-Shivakumar D et al. (2004), Proc Natl Acad Sci U S A, 101, 14379-14384. Molecular dynamics studies of ground state and intermediate of the hyperthermophilic indole-3-glycerol phosphate synthase. DOI:10.1073/pnas.0406002101. PMID:15452341.
- Hennig M et al. (2002), J Mol Biol, 319, 757-766. The Catalytic Mechanism of Indole-3-glycerol Phosphate Synthase: Crystal Structures of Complexes of the Enzyme from Sulfolobus solfataricus with Substrate Analogue, Substrate, and Product. DOI:10.1016/s0022-2836(02)00378-9. PMID:12054868.
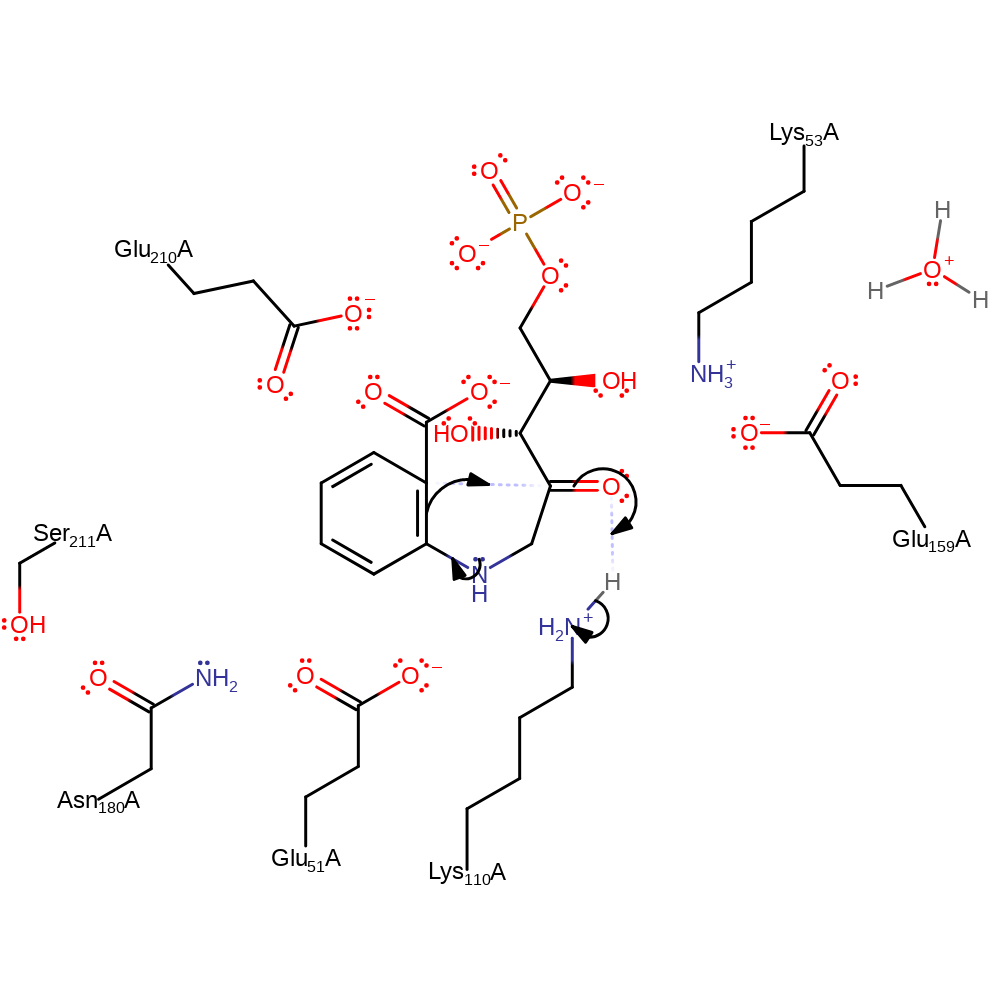
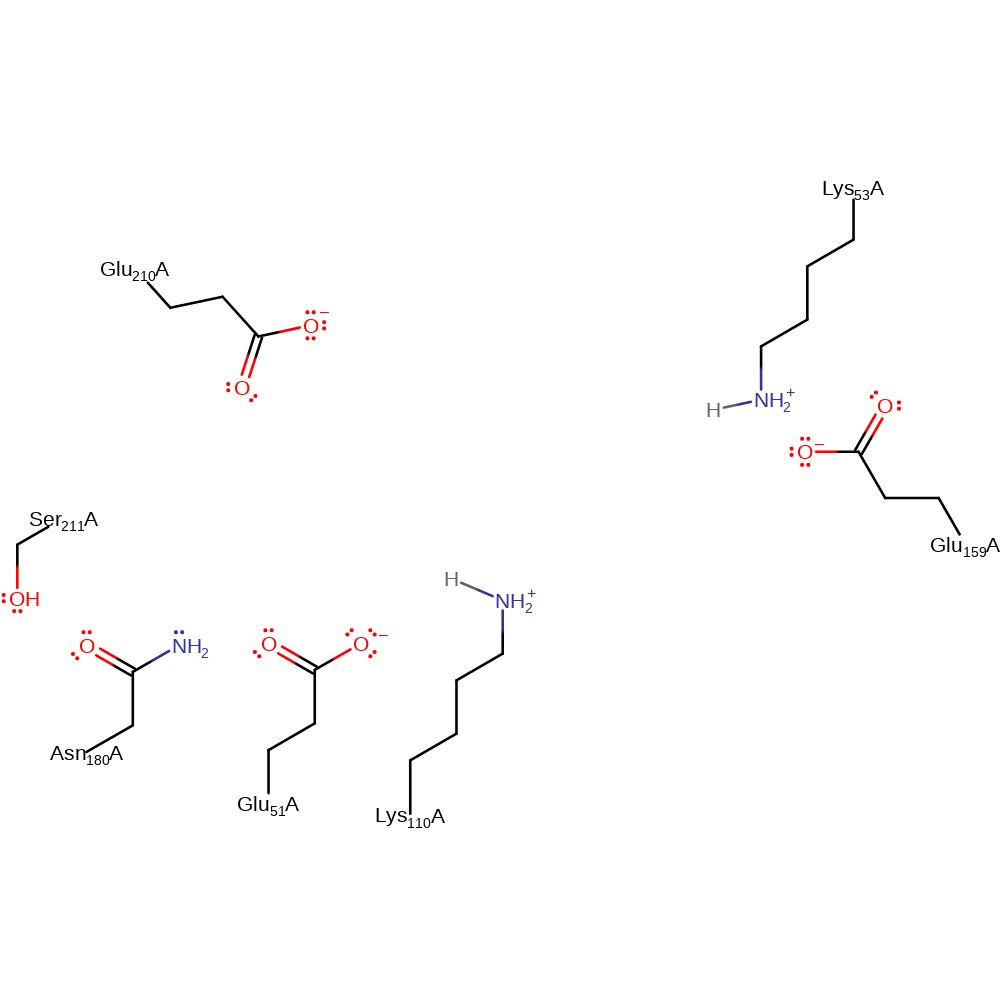
Step 1. The nitrogen in the substrate initiates a double bond rearrangement which results in the cyclisation of the intermediate, and deprotonation of Lys110.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys53A | electrostatic stabiliser, hydrogen bond donor |
| Glu51A | hydrogen bond acceptor, electrostatic stabiliser, increase acidity |
| Lys110A | hydrogen bond donor |
| Glu159A | hydrogen bond acceptor, increase nucleophilicity |
| Asn180A | hydrogen bond donor, hydrogen bond acceptor, electrostatic stabiliser |
| Ser211A | hydrogen bond donor, electrostatic stabiliser |
| Glu210A | electrostatic stabiliser |
| Lys110A | proton donor |
Chemical Components
ingold: intramolecular nucleophilic addition, proton transfer, cyclisation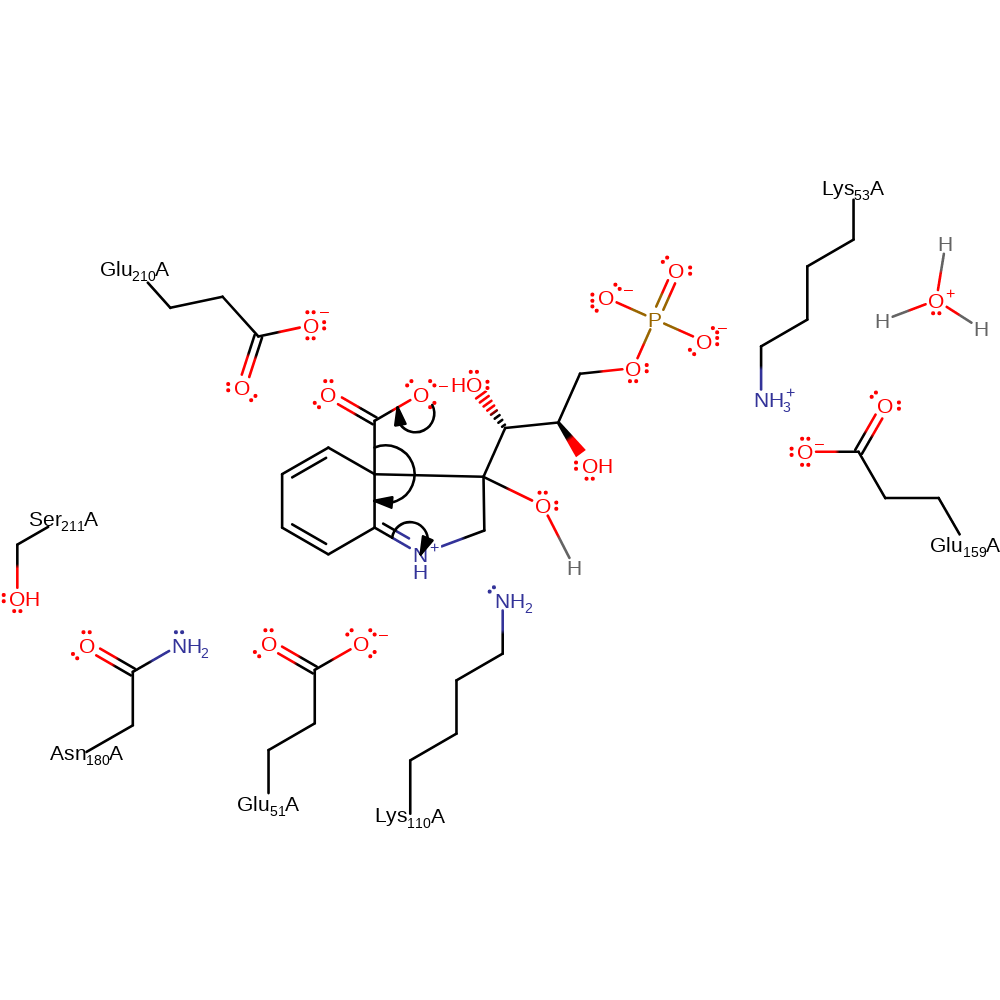
Step 2. The carboxylic acid group undergoes elimination from the substrate resulting indecarboxylation.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys53A | electrostatic stabiliser, hydrogen bond donor |
| Glu51A | hydrogen bond acceptor, electrostatic stabiliser |
| Lys110A | hydrogen bond donor |
| Glu159A | hydrogen bond acceptor |
| Asn180A | hydrogen bond donor, hydrogen bond acceptor, electrostatic stabiliser |
| Ser211A | hydrogen bond donor, electrostatic stabiliser |
| Glu210A | electrostatic stabiliser |
Chemical Components
ingold: unimolecular elimination by the conjugate base, decarboxylation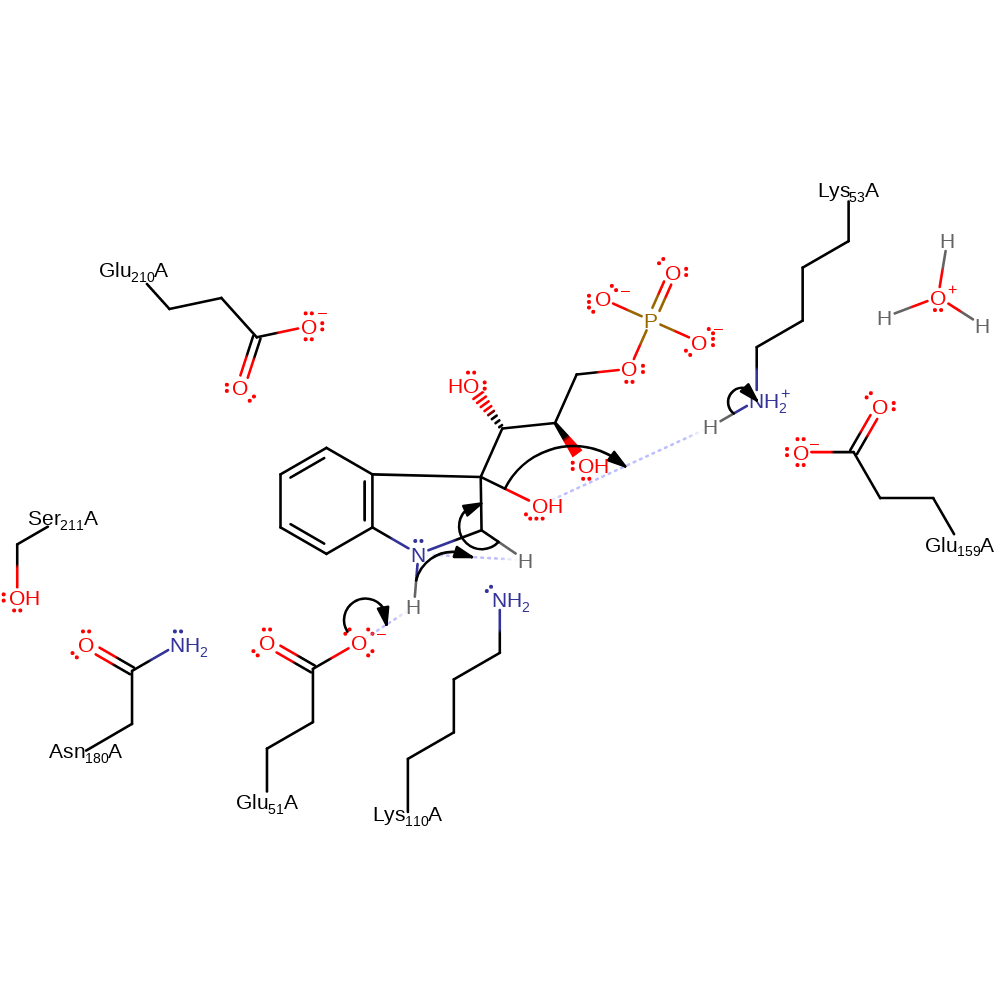
Step 3. Glu51 deprotonates the newly formed ring nitrogen, which in turn abstracts a proton from the adjacent CH2, eliminating water with concomitant deprotonation of Lys53.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys53A | electrostatic stabiliser, hydrogen bond donor |
| Glu51A | increase basicity, hydrogen bond acceptor, electrostatic stabiliser |
| Lys110A | hydrogen bond acceptor |
| Glu159A | hydrogen bond acceptor |
| Asn180A | hydrogen bond donor, hydrogen bond acceptor, electrostatic stabiliser |
| Ser211A | hydrogen bond donor, electrostatic stabiliser |
| Glu210A | electrostatic stabiliser |
| Glu159A | activator |
| Glu51A | proton acceptor |
| Lys53A | proton donor |
Chemical Components
proton transfer, ingold: intramolecular elimination, dehydration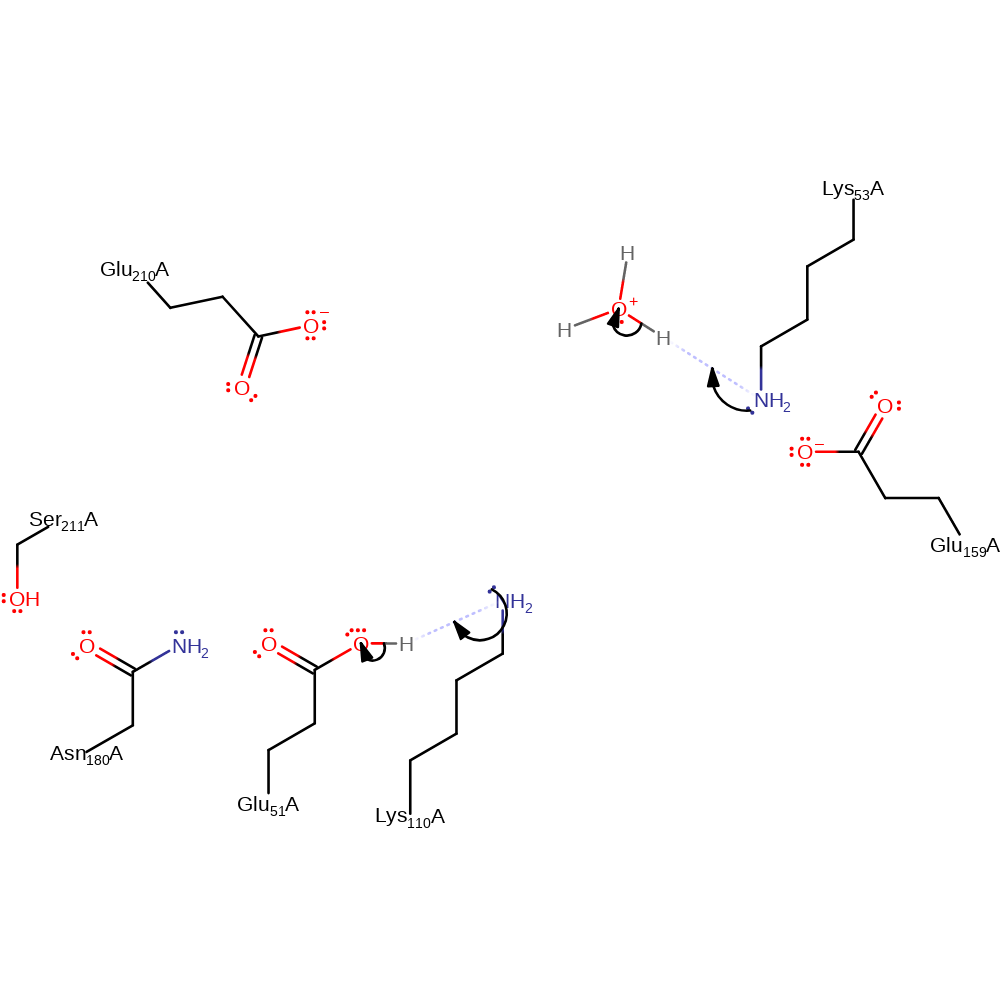
Step 4. In this inferred return step, Lys53 accepts a proton (probably from bulk solvent) and Lys110 abstracts the proton from Glu51.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu51A | hydrogen bond acceptor, electrostatic stabiliser |
| Lys110A | hydrogen bond donor |
| Glu159A | hydrogen bond acceptor |
| Asn180A | hydrogen bond donor, hydrogen bond acceptor |
| Ser211A | hydrogen bond donor, electrostatic stabiliser |
| Glu159A | activator |
| Asn180A | increase acidity |
| Glu51A | proton donor |
| Lys110A | proton acceptor |
| Lys53A | proton acceptor |





 Download:
Download: