Beta-lactamase (Class D)
Class D beta lactamases are able to break down oxacillin and cloxacillin thus playing a vital role in antibiotic resistance in bacteria as they can prevent these compounds from inhibiting the DD peptidases involved in bacterial cell wall synthesis. As part of the family of serine beta lactamases, which also includes classes A B and C of the beta lactamases, they share significant homology with the DD peptidases, indicating that they diverged from a common ancestor. However, they are able to hydrolyse the acyl enzyme intermediate that forms through initial nucleophilic attack on the beta-lactam ring much more quickly, thus do not become inhibited by the beta lactams in the same way.
Reference Protein and Structure
- Sequence
-
P13661
 (3.5.2.6)
(3.5.2.6)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli (Bacteria)

- PDB
-
1m6k
- Structure of the OXA-1 class D beta-lactamase
(1.5 Å)



- Catalytic CATH Domains
-
3.40.710.10
 (see all for 1m6k)
(see all for 1m6k)
Enzyme Reaction (EC:3.5.2.6)
Enzyme Mechanism
Introduction
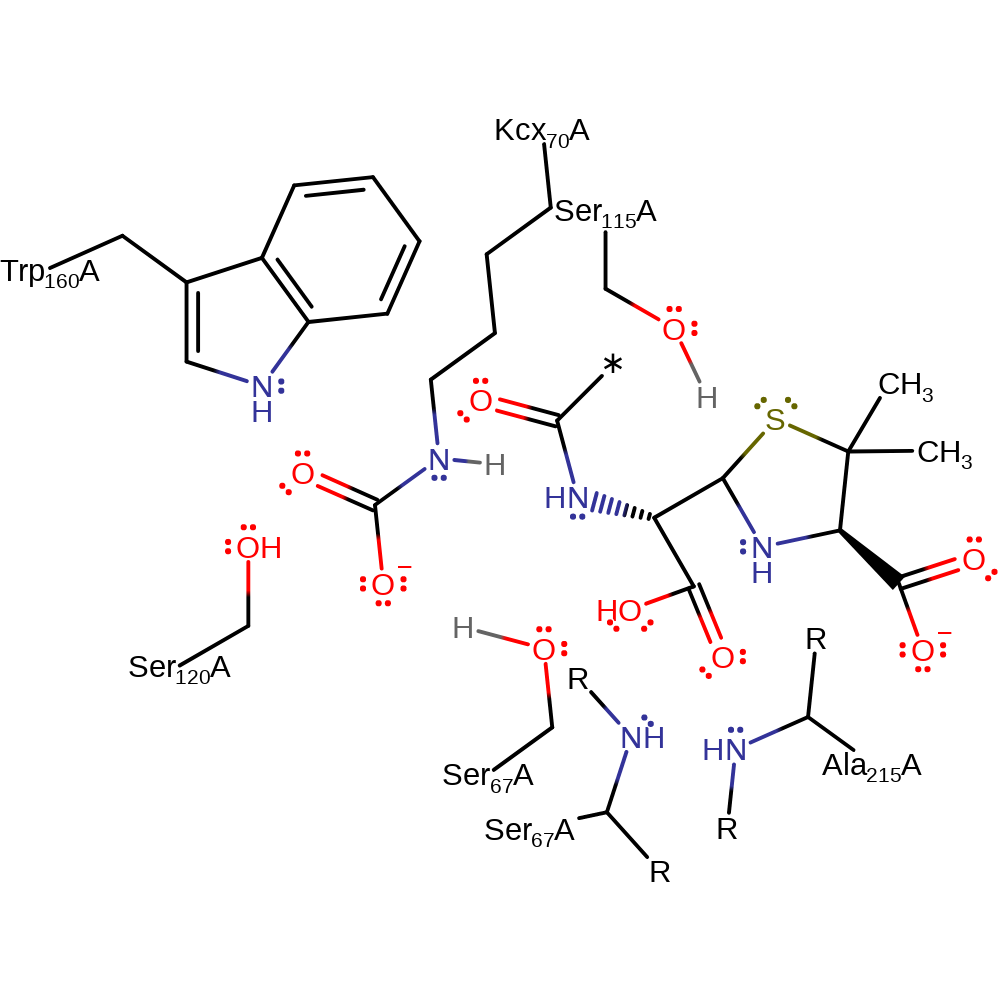
Ser 67, having been deprotonated by carboxylated Lys 70, is able to act as a nucleophile to attack the strained peptide bond in the beta lactam ring and form a tetrahedral intermediate, stabilised by the backbone amide of Ser 67. The collapse of this intermediate, facilitated by protonation of the leaving group by the epsilon NH of the carboxylated Lys 70, results in an acyl enzyme intermediate. This can be hydrolysed because a water molecule, activated by deprotonation by Lys 70, can attack the intermediate, resulting in the formation of a carboxylic acid and the regeneration of the catalytically active Ser 67 residue.
Catalytic Residues Roles
| UniProt | PDB* (1m6k) | ||
| Ser71 (main-N) | Ser67(46)A (main-N) | Acts as nucleophile to attack the beta lactam ring, forming an oxyanion intermediate which is stabilised by a hydrogen bond between its main chain amide and the oxyanion. Subsequent collapse and hydrolysis of the intermediates results in product formation. | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| Ala218 (main-N), Ser71 (main-N) | Ala215(193)A (main-N), Ser67(46)A (main-N) | Form the oxyanion hole. | hydrogen bond donor, electrostatic stabiliser |
| Ser118 | Ser115(93)A | Acts as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, proton relay |
| Trp163, Ser123 | Trp160(138)A, Ser120(98)A | Act to stabilise and activate the carbamylated lysine residue. | hydrogen bond donor, electrostatic stabiliser |
| Ser71 | Ser67(46)A | Acts as a catalytic nuclephile and is activated by the carbamylated lysine residue. | covalently attached, nucleofuge, nucleophile, proton acceptor, proton donor |
| Lys74 (ptm) | Kcx70(49)A (ptm) | This residue is post-translationally carbamylated and acts as a general acid/base. | proton relay, proton acceptor, proton donor |
Chemical Components
proton transfer, bimolecular nucleophilic addition, intermediate formation, enzyme-substrate complex formation, overall reactant used, unimolecular elimination by the conjugate base, decyclisation, proton relay, enzyme-substrate complex cleavage, intermediate collapse, intermediate terminated, overall product formed, native state of enzyme regeneratedReferences
- Maveyraud L et al. (2000), Structure, 8, 1289-1298. Insights into Class D β-Lactamases Are Revealed by the Crystal Structure of the OXA10 Enzyme from Pseudomonas aeruginosa. DOI:10.1016/s0969-2126(00)00534-7. PMID:11188693.
- Leonard DA et al. (2013), Acc Chem Res, 46, 2407-2415. Class D β-lactamases: a reappraisal after five decades. DOI:10.1021/ar300327a. PMID:23902256.
- Sun T et al. (2003), Protein Sci, 12, 82-91. Comparison of -lactamases of classes A and D: 1.5-A crystallographic structure of the class D OXA-1 oxacillinase. DOI:10.1110/ps.0224303. PMID:12493831.
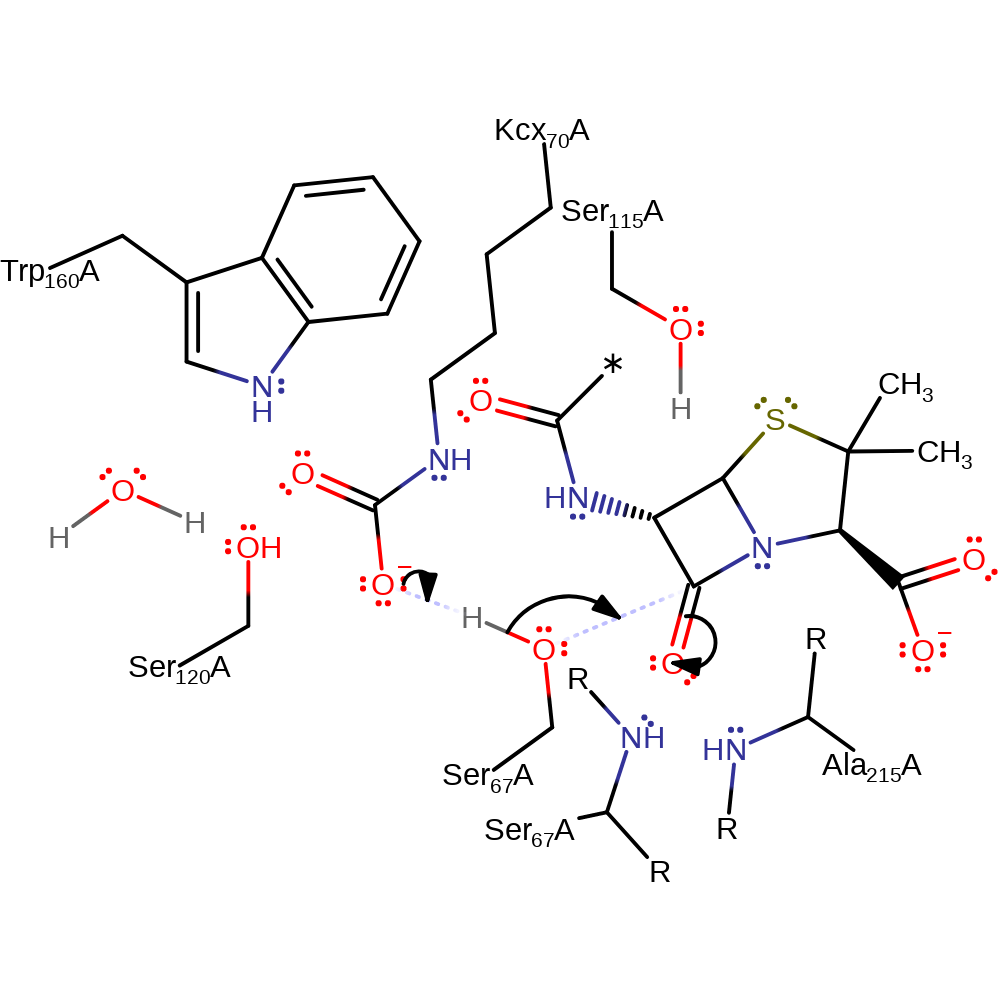
Step 1. Carbamylated lysine (Kcx70) deprotonates the alcohol of Ser67, which initiates a nucleophilic addition to the carbonyl group of the betalactam ring.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser115(93)A | hydrogen bond donor, hydrogen bond acceptor |
| Ala215(193)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Trp160(138)A | hydrogen bond donor, electrostatic stabiliser |
| Ser67(46)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Ser120(98)A | hydrogen bond donor, electrostatic stabiliser |
| Ser67(46)A | proton donor, nucleophile |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, enzyme-substrate complex formation, overall reactant used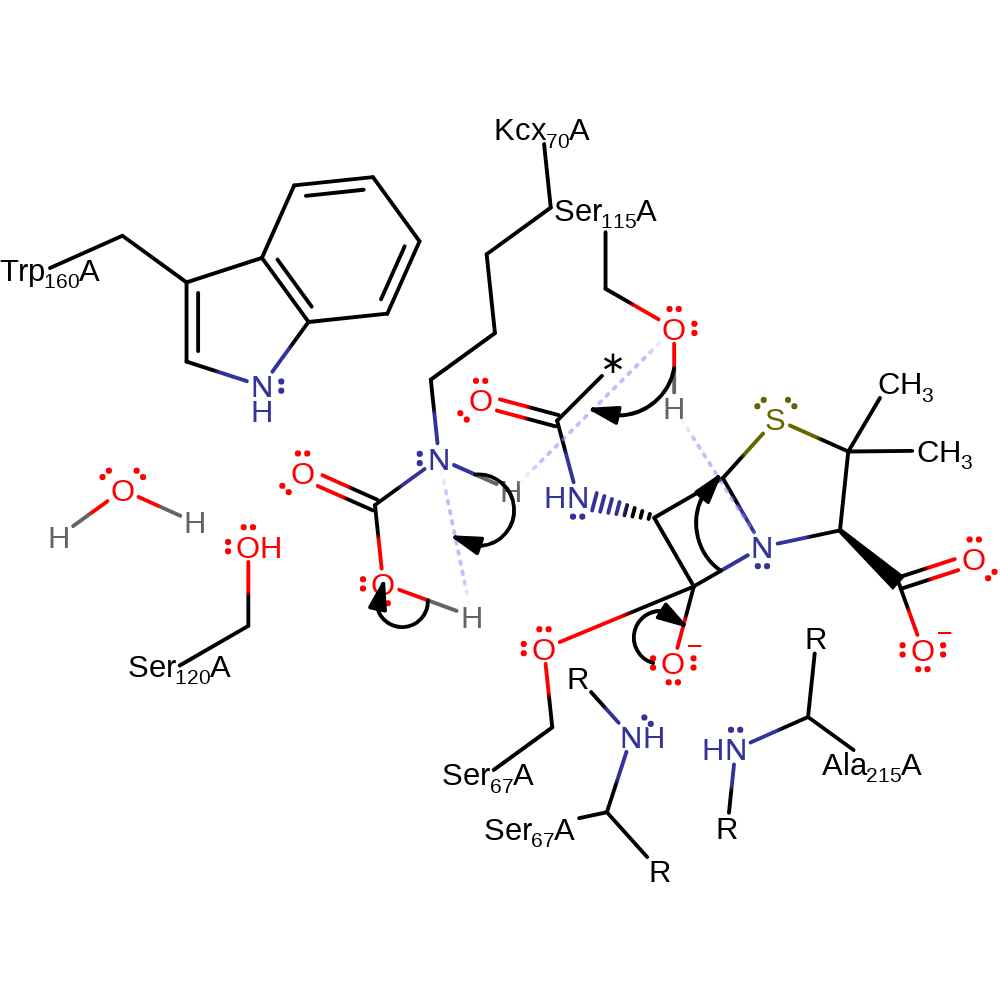
Step 2. The tetrahedral intermediate collapses, cleaving the C-N bond, which deprotonates Ser115, which in turn deprotonates the secondary amine of Kcx70, which deprotonates the carboxyl group of Kcx70.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser115(93)A | hydrogen bond donor, hydrogen bond acceptor, proton relay |
| Ala215(193)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Trp160(138)A | hydrogen bond donor, electrostatic stabiliser |
| Ser67(46)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Ser120(98)A | hydrogen bond donor, electrostatic stabiliser |
| Ser67(46)A | covalently attached |
| Kcx70(49)A (ptm) | proton donor |
| Ser115(93)A | proton acceptor, proton donor |
| Kcx70(49)A (ptm) | proton acceptor, proton relay |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, intermediate formation, decyclisation, proton relay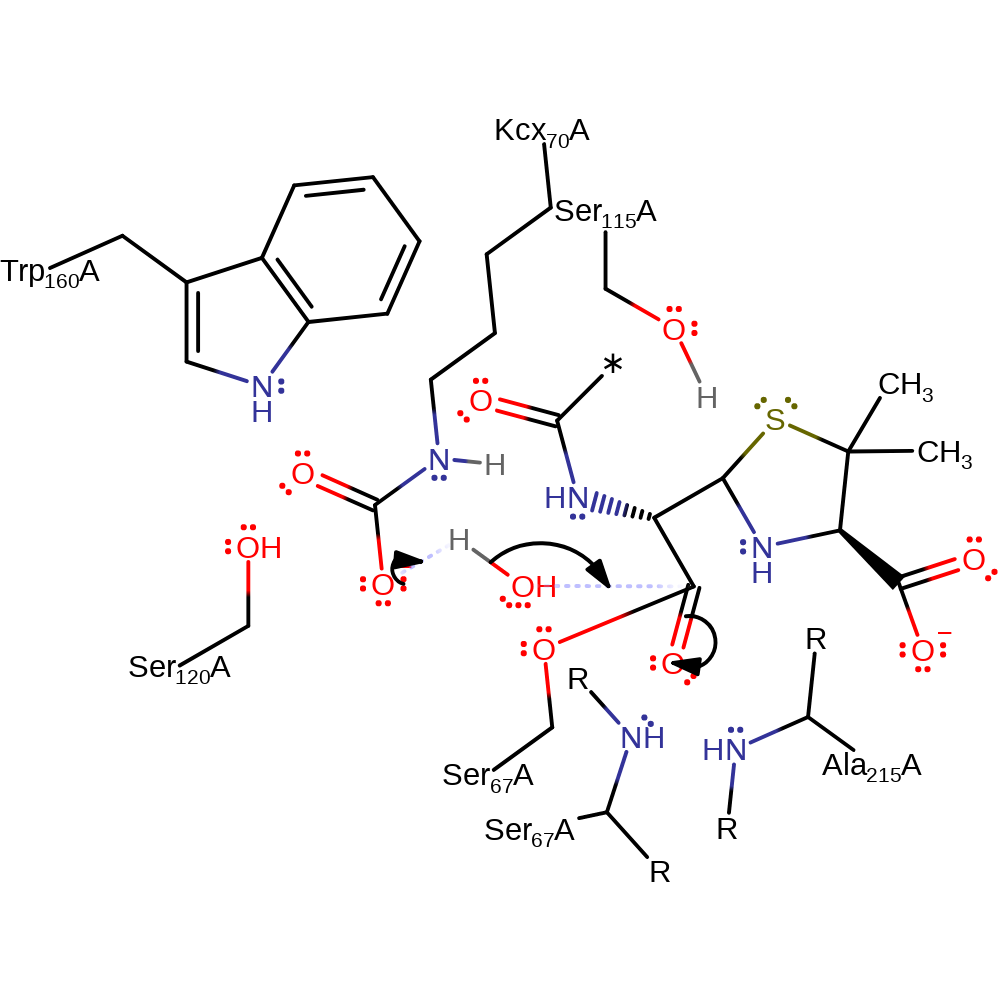
Step 3. Kcx70 deprotonates water, which attacks the carbonyl carbon of the covalently attached intermediate in a nucleophilic addition.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser115(93)A | hydrogen bond donor, hydrogen bond acceptor |
| Ala215(193)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Trp160(138)A | hydrogen bond donor, electrostatic stabiliser |
| Ser67(46)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Ser120(98)A | hydrogen bond donor, electrostatic stabiliser |
| Ser67(46)A | covalently attached |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used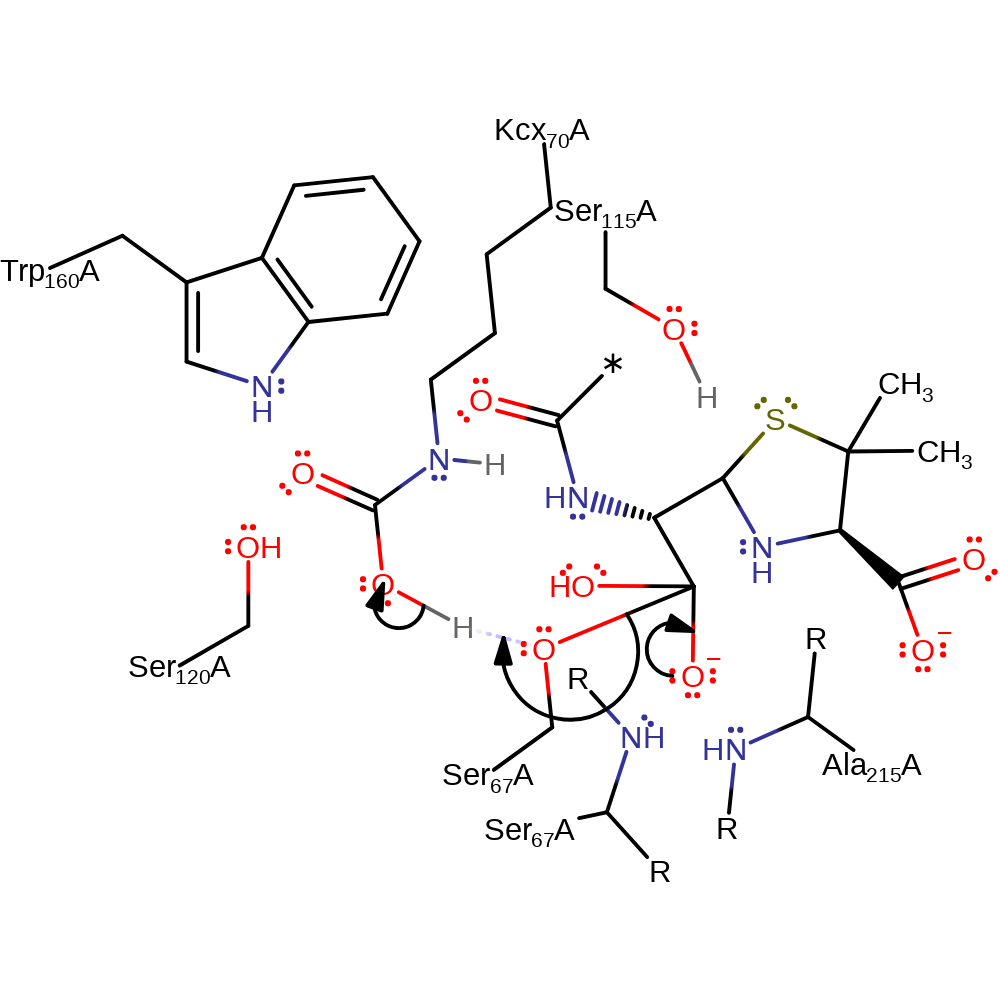
Step 4. The tetrahedral intermediate collapses, eliminating Ser67, which reprotonates from Kcx70, producing the product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser115(93)A | hydrogen bond donor, hydrogen bond acceptor |
| Ala215(193)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Trp160(138)A | hydrogen bond donor, electrostatic stabiliser |
| Ser67(46)A (main-N) | hydrogen bond donor, electrostatic stabiliser, hydrogen bond acceptor |
| Ser120(98)A | hydrogen bond donor, electrostatic stabiliser |
| Kcx70(49)A (ptm) | proton donor |
| Ser67(46)A | proton acceptor, nucleofuge |



 Download:
Download: