Hydroxyacylglutathione hydrolase
Glyoxalase II participates in the cellular detoxification of cytotoxic and mutagenic 2-oxoaldehydes. Glyoxalase I converts methylglyoxal (a cytotoxic byproduct of many cellular reactions) and glutathione into S-D-lactoylglutathione (SLG) which is also cytotoxic; the role of glyoxalase II is to hydrolyse SLG into D-lactic acid and glutathione.
The substrate of this reaction is the product of Lactoylglutathione lyase, an enzyme which catalyses the initial steps in detoxifying methyl-glyoxyl. This enzyme regnerates the glutathione cofactor utilised by lactoylglutathione lyase.
Reference Protein and Structure
- Sequence
-
Q16775
 (3.1.2.6)
(3.1.2.6)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
1qh5
- HUMAN GLYOXALASE II WITH S-(N-HYDROXY-N-BROMOPHENYLCARBAMOYL)GLUTATHIONE
(1.45 Å)



- Catalytic CATH Domains
-
3.60.15.10
 (see all for 1qh5)
(see all for 1qh5)
- Cofactors
- Zinc(2+) (2) Metal MACiE
Enzyme Reaction (EC:3.1.2.6)
Enzyme Mechanism
Introduction
Glyoxalase II has a binuclear centre that has affinity for both Zn(II) and Fe(II) and is functional with either metal in either of its metal-binding sites. Both metals coordinate a bridging hydroxide ion. This hydroxide nucleophile attacks the carbonyl carbon of the thioester group of SLG. A tetrahedral intermediate is formed. Metal 1 stabilises the negative charge forming on the thioester oxygen, and metal 2 stabilises the negative charge forming on the thioester sulphur (thus making the thiolate anion a better leaving group). The tetrahedral intermediate collapses. Lactic acid and glutathione (still bound to metal 2 as a thiolate) are formed. Asp58 accepts a proton from the bridging oxygen to generate the lactate, this proton is then transferred to the thiolate to form glutathione. The products are released and new water bridges the zinc ions, the water is deprotonated by Asp58 regenerating the active site.
Catalytic Residues Roles
| UniProt | PDB* (1qh5) | ||
| Asp106 | Asp58A | Deprotonates lactic acid and transfers the proton to reform glutathione. Also deprotonates a water molecule to regenerate the active site. | hydrogen bond acceptor, metal ligand, proton acceptor, proton donor, activator, electrostatic stabiliser |
| Asp182 | Asp134A | Acts as a bridging ligand between the two zinc sites. | metal ligand |
| His102, His104, His158 | His54A, His56A, His110A | Forms part of the zinc 1 binding site. | metal ligand |
| His107, His221 | His59A, His173A | Forms part of the zinc 2 binding site. | metal ligand |
Chemical Components
overall reactant used, bimolecular nucleophilic addition, unimolecular elimination by the conjugate base, overall product formed, proton transfer, native state of enzyme regenerated, native state of cofactor regeneratedReferences
- Zang TM et al. (2001), J Biol Chem, 276, 4788-4795. Arabidopsis Glyoxalase II Contains a Zinc/Iron Binuclear Metal Center That Is Essential for Substrate Binding and Catalysis. DOI:10.1074/jbc.m005090200. PMID:11085979.
- Chen SL et al. (2009), J Inorg Biochem, 103, 274-281. Reaction mechanism of the binuclear zinc enzyme glyoxalase II - A theoretical study. DOI:10.1016/j.jinorgbio.2008.10.016. PMID:19062100.
- Cameron AD et al. (1999), Structure, 7, 1067-1078. Crystal structure of human glyoxalase II and its complex with a glutathione thiolester substrate analogue. DOI:10.1016/s0969-2126(99)80174-9. PMID:10508780.

Step 1. Zinc activate water attacked the carbonyl carbon of the (R)-S-lactoylglutathione substrate in a nucleophilic addition, forming a tetrahedral intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His54A | metal ligand |
| His56A | metal ligand |
| His110A | metal ligand |
| Asp58A | hydrogen bond acceptor, metal ligand, electrostatic stabiliser |
| Asp134A | metal ligand |
| His59A | metal ligand |
| His173A | metal ligand |
Chemical Components
overall reactant used, ingold: bimolecular nucleophilic addition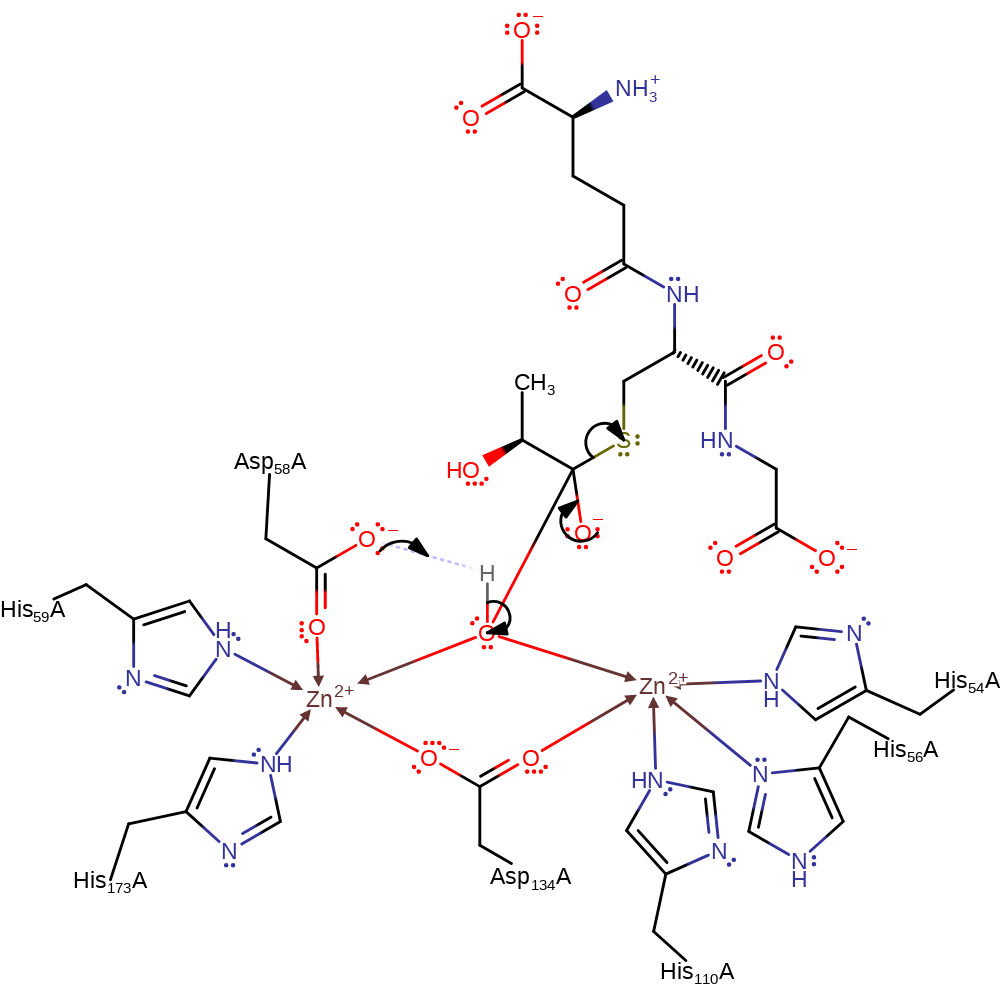
Step 2. The tetrahedral intermediate collapses releasing the (R)-lactate product and the thiolate form of glutathione. Asp58 accepts a proton from the bridging oxygen.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His54A | metal ligand |
| His56A | metal ligand |
| Asp58A | metal ligand |
| His59A | metal ligand |
| His110A | metal ligand |
| Asp134A | metal ligand |
| His173A | metal ligand |
| Asp58A | hydrogen bond acceptor, electrostatic stabiliser |
| Asp58A | proton acceptor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, overall product formed, proton transfer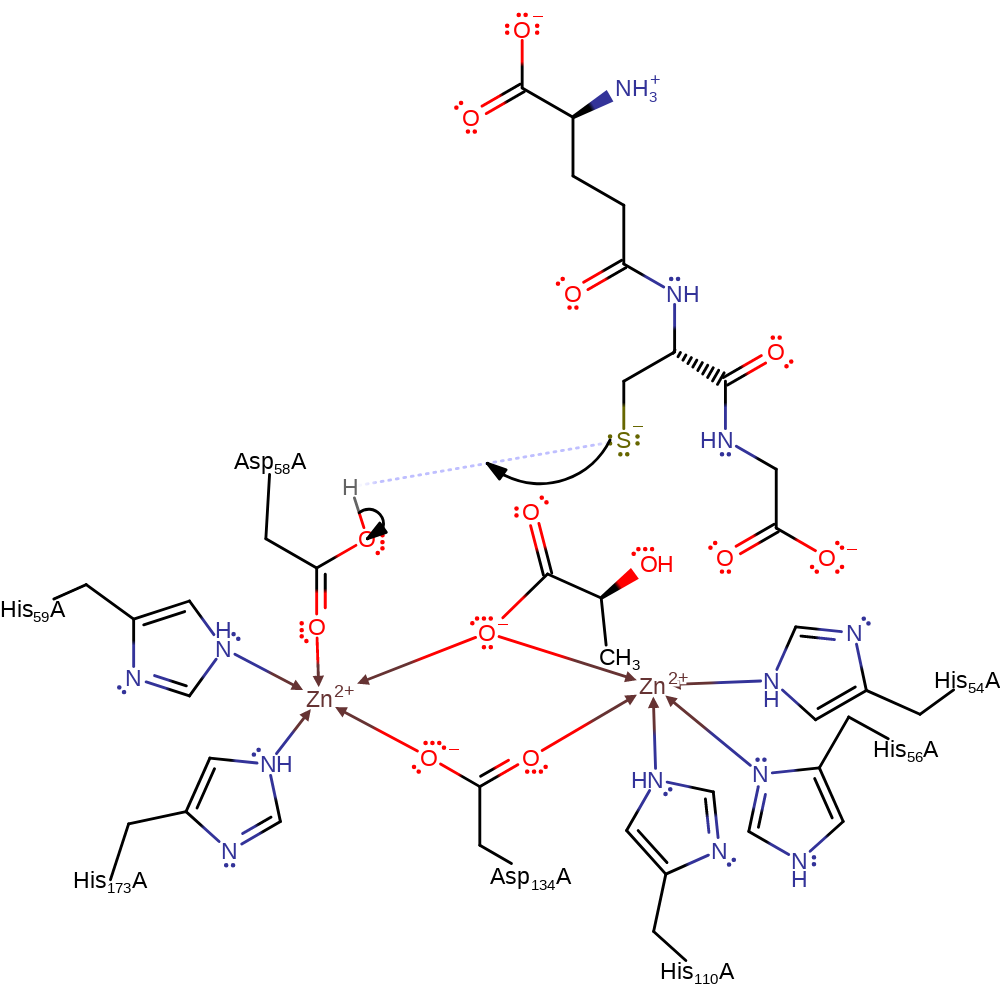
Step 3. The thiolate of glutathione deprotonates Asp58, regenerating the glutathione cofactor used in the initial formation of hydroacetylglutathione.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp58A | hydrogen bond acceptor, activator |
| His54A | metal ligand |
| His56A | metal ligand |
| His110A | metal ligand |
| Asp134A | metal ligand |
| Asp58A | metal ligand |
| His59A | metal ligand |
| His173A | metal ligand |
| Asp58A | proton donor |
Chemical Components
proton transfer, native state of enzyme regenerated
Step 4. Asp58 deprotonates a new bridging water molecule regenerating the active site. This proton can then be transferred from Asp58 to solution.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His54A | metal ligand |
| His56A | metal ligand |
| Asp58A | metal ligand |
| His59A | metal ligand |
| His110A | metal ligand |
| Asp134A | metal ligand |
| His173A | metal ligand |
| Asp58A | proton acceptor |





 Download:
Download: