Xanthine dehydrogenase (mammalian)
Mammalian xanthine oxidoreductase catalyses the hydroxylation of hypoxanthine and xanthine, the last two stages in the biosynthesis pathway of urate. The enzyme is first formed as xanthine dehydrogenase (XDH), and exists mostly in this form within the cell but it can be readily converted to the oxidase form xanthine oxidase (XO) by oxidation of sulfhydryl residues or by proteolysis.
The active form of the enzyme is a homodimer of a three chain subunits, where both units contain one molybdenum centre, one flavin (FAD) centre and two Fe-S clusters. The oxidation of xanthine to uric acid takes place at the molybdenum centre and results in a two electron reduction of the metal from Mo(VI) to Mo(IV). The enzyme is subsequently reoxidised by molecular oxygen in a reaction that occurs at the FAD cofactor.
Xanthine dehydrogenase (EC 1.17.1.4) and xanthine oxidase (EC 1.17.3.2 - M-CSA ID:987) are two variants of the same gene product. The former prefers NAD+ as the oxidising substrate whereas the latter uses dioxygen exclusively.Reference Protein and Structure
- Sequence
-
P80457
 (1.17.1.4, 1.17.3.2)
(1.17.1.4, 1.17.3.2)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Bos taurus (Cattle)

- PDB
-
1v97
- Crystal Structure of Bovine Milk Xanthine Dehydrogenase FYX-051 bound form
(1.94 Å)



- Catalytic CATH Domains
-
3.30.365.10
 (see all for 1v97)
(see all for 1v97)
- Cofactors
- Di-mu-sulfido-diiron(2+) (2), Molybdopterin (1), Dioxothiomolybdenum(vi) ion (1), Fadh2(2-) (1) Metal MACiE
Enzyme Reaction (EC:1.17.1.4)
Enzyme Mechanism
Introduction
The most recently proposed mechanism describes a catalytically labile Mo-OH group of the oxidised Mo(VI) enzyme initiating catalysis by base assisted nucleophilic attack on the carbon of the centre to be hydroxylated, with concomitant hydride transfer. This yields a reduced Mo(IV)-SH, derived from the Mo(VI)=S of the oxidised enzyme, with the product remaining coordinated to the molybdenum via the newly introduced hydroxyl group. The product is displaced by attack of a water molecule at the Mo centre. Glu-1261 is well positioned to play the role of the general base in abstracting a proton from the Mo-OH group, while Arg880 stabilises the anionic charge on the substrate while Glu802 is implicated in stabilising the desired product tautomer.
The mechanism shown here strongly supports all the available evidence. The pH dependence of the reaction strongly suggests that the active site base (Glu1261A) is initially in its unprotonated state and that the enzyme only works on the substrate in its neutral state [PMID:15134930]. The mechanism shown in here is supported by mutational spectroscopic computational and crystallographic studies [PMID:15148401, PMID:15581570, PMID:15134930, PMID:15265866]. There has been some debate as to whether the catalytically labile oxygen is from the Mo=O or Mo-OH groups. Isotope labelling experiments of solvent shows that the catalytically labile group is the Mo-OH and that it is replenished from solvent at the end of the catalytic cycle [PMID:15148401, PMID:15581570].
Catalytic Residues Roles
| UniProt | PDB* (1v97) | ||
| Glu802 | Glu802A | The residue is thought to influence the tautomerisation step, relaying a proton from Glu1261 to a nitrogen on the substrate by lowering the energy of the desired tautomer, with concomitant hydride transfer to the metal cofactor. | hydrogen bond acceptor, electrostatic stabiliser |
| Arg880 | Arg880A | The positively charged gaunidinium side chain acts to stabilise the build up of negative charge on the substrate once nucleophilic attack has occurred. | hydrogen bond donor, electrostatic stabiliser |
| Glu1261 | Glu1261A | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic substitution, hydride transfer, bimolecular nucleophilic addition, overall reactant used, cofactor used, intermediate formation, redox reaction, radical formation, native state of cofactor regenerated, overall product formed, electron relay, electron transfer, radical termination, coordination to a metal ion, decoordination from a metal ion, intermediate terminated, aromatic unimolecular elimination by the conjugate base, aromatic bimolecular nucleophilic addition, native state of enzyme regeneratedReferences
- Choi EY et al. (2004), J Inorg Biochem, 98, 841-848. Studies on the mechanism of action of xanthine oxidase. DOI:10.1016/j.jinorgbio.2003.11.010. PMID:15134930.
- Reschke S et al. (2017), Inorg Chem, 56, 2165-2176. Protonation and Sulfido versus Oxo Ligation Changes at the Molybdenum Cofactor in Xanthine Dehydrogenase (XDH) Variants Studied by X-ray Absorption Spectroscopy. DOI:10.1021/acs.inorgchem.6b02846. PMID:28170236.
- Cao H et al. (2014), Biochemistry, 53, 533-541. Substrate orientation and specificity in xanthine oxidase: crystal structures of the enzyme in complex with indole-3-acetaldehyde and guanine. DOI:10.1021/bi401465u. PMID:24397336.
- Metz S et al. (2010), J Phys Chem B, 114, 1506-1517. QM/MM studies of xanthine oxidase: variations of cofactor, substrate, and active-site Glu802. DOI:10.1021/jp909999s. PMID:20050623.
- Pauff JM et al. (2009), J Biol Chem, 284, 8760-8767. Substrate Orientation and Catalysis at the Molybdenum Site in Xanthine Oxidase: CRYSTAL STRUCTURES IN COMPLEX WITH XANTHINE AND LUMAZINE. DOI:10.1074/jbc.m804517200. PMID:19109252.
- Metz S et al. (2009), J Am Chem Soc, 131, 14885-14902. A combined QM/MM study on the reductive half-reaction of xanthine oxidase: substrate orientation and mechanism. DOI:10.1021/ja9045394. PMID:19788181.
- Hille R (2005), Arch Biochem Biophys, 433, 107-116. Molybdenum-containing hydroxylases. DOI:10.1016/j.abb.2004.08.012. PMID:15581570.
- Zhang XH et al. (2005), Inorg Chem, 44, 1466-1471. A Theoretical Study on the Mechanism of the Reductive Half-Reaction of Xanthine Oxidase. DOI:10.1021/ic048730l. PMID:15732988.
- Okamoto K et al. (2004), Proc Natl Acad Sci U S A, 101, 7931-7936. The crystal structure of xanthine oxidoreductase during catalysis: Implications for reaction mechanism and enzyme inhibition. DOI:10.1073/pnas.0400973101. PMID:15148401.
- Leimkühler S et al. (2004), J Biol Chem, 279, 40437-40444. The Role of Active Site Glutamate Residues in Catalysis ofRhodobacter capsulatusXanthine Dehydrogenase. DOI:10.1074/jbc.m405778200. PMID:15265866.
- Stockert AL et al. (2002), J Am Chem Soc, 124, 14554-14555. The Reaction Mechanism of Xanthine Oxidase: Evidence for Two-Electron Chemistry Rather Than Sequential One-Electron Steps. DOI:10.1021/ja027388d.
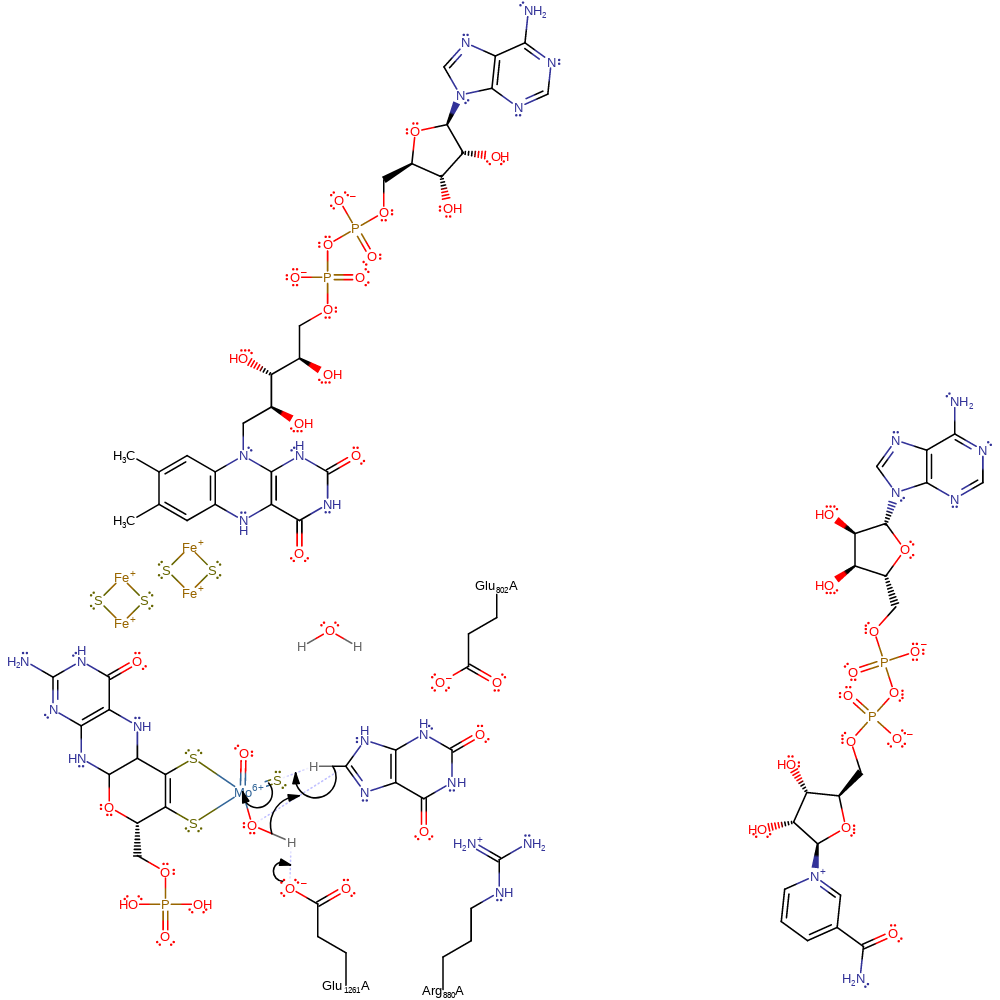
Step 1. Glu1261 deprotonates the MTE bound hydroxide, activating it for nucleophilic attack upon the xanthine substrate, resulting in a nucleophilic addition of the substrate to the cofactor. Xanthine then transfers a hydride to the sulfur bound to the molybdenum ion (Mo=S) resulting in a two electron reduction of the Mo(VI) to Mo(IV) and a thiol bound to molybdenum.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu1261A | hydrogen bond acceptor, electrostatic stabiliser |
| Arg880A | hydrogen bond donor, electrostatic stabiliser |
| Glu802A | hydrogen bond acceptor, electrostatic stabiliser |
| Glu1261A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, hydride transfer, ingold: bimolecular nucleophilic addition, overall reactant used, cofactor used, intermediate formation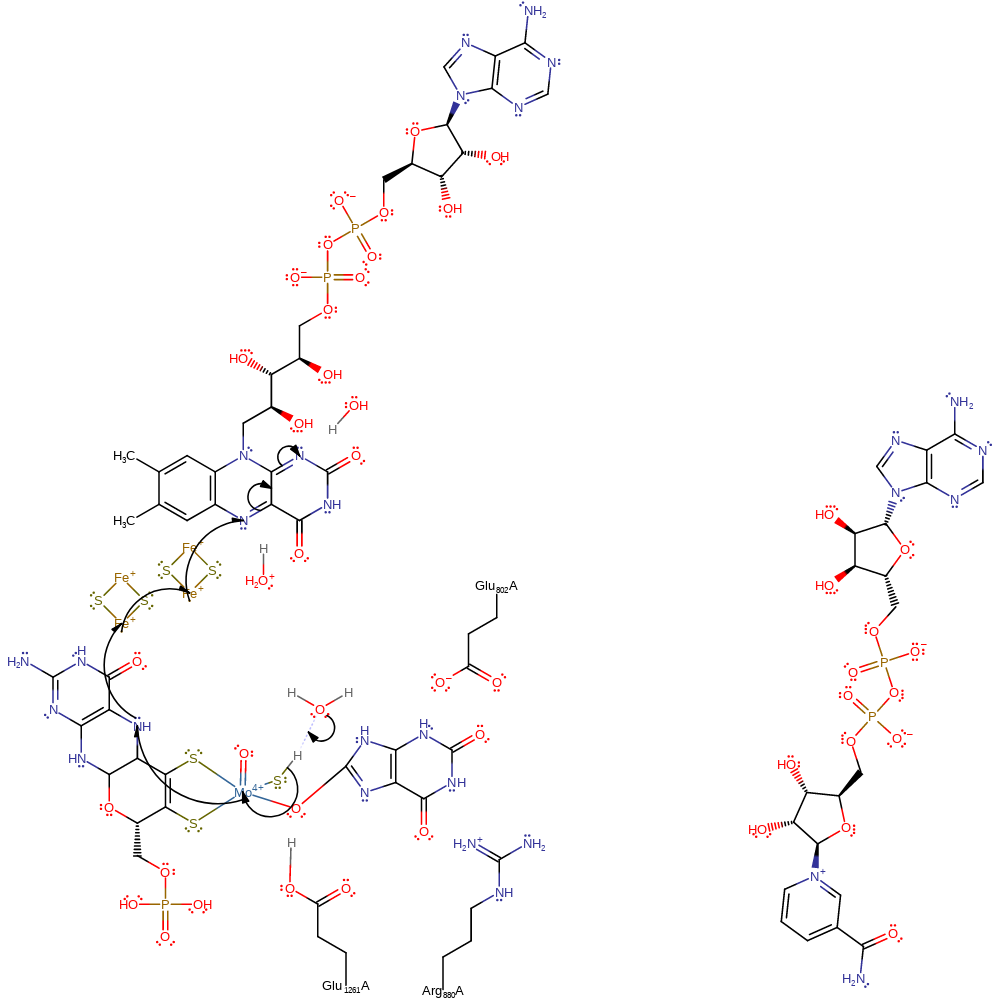
Step 2. Water deprotonates the molybdenum bound thiol, reforming the Mo=S species and a single electron transfer from the Mo(IV) through the rest of the cofactor and two iron-sulfur clusters to a bound FAD cofactor, forming Mo(V) and a radical on the FAD cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu1261A | hydrogen bond donor, electrostatic stabiliser |
| Arg880A | hydrogen bond donor |
| Glu802A | hydrogen bond acceptor |
Chemical Components
proton transfer, redox reaction, radical formation, cofactor used, native state of cofactor regenerated, intermediate formation, overall product formed, electron relay
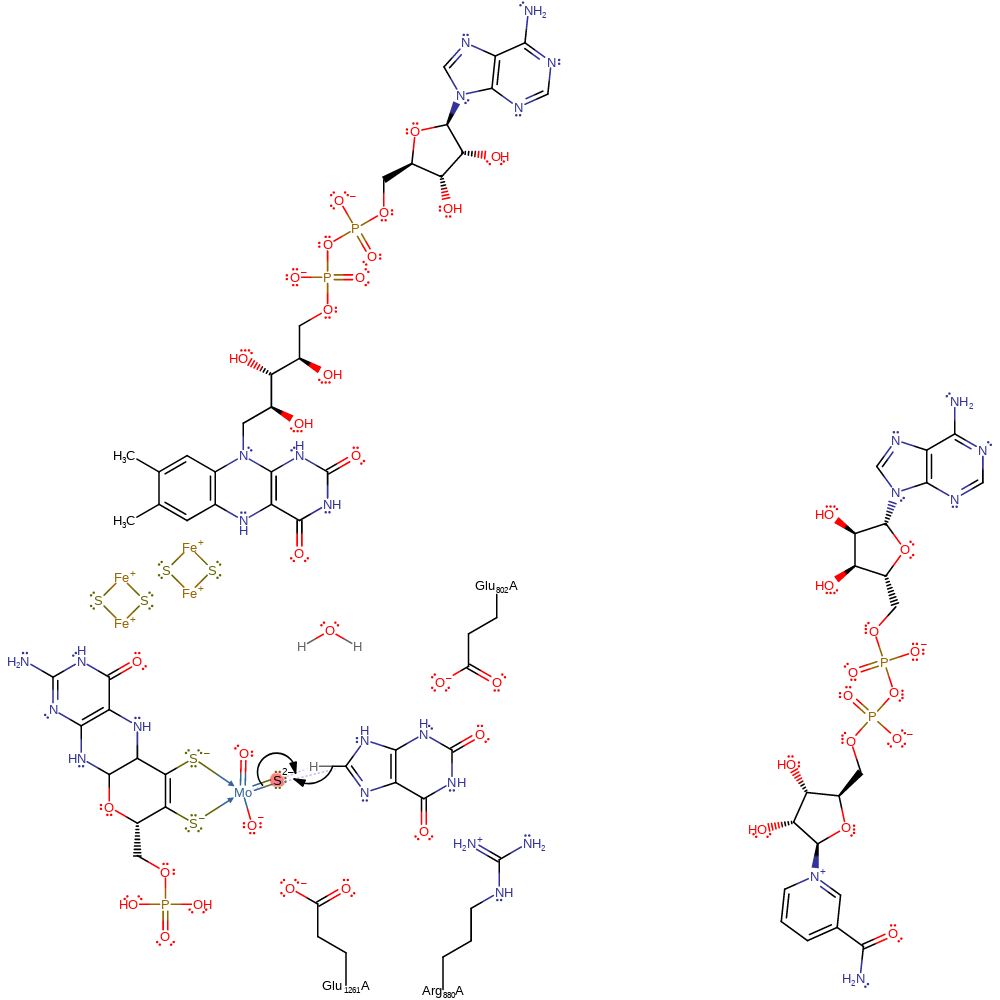
Step 3. A free hydroxide ion attacks the Mo(V), releasing the urate product (oxidised xanthine), which is re-protonated from the Glu1261, in a nucleophilic substitution reaction. This also results in the second electron transfer from the Mo(V) through the rest of the cofactor and two iron-sulfur clusters to a bound FAD cofactor, which then deprotonates a hydroxonium ion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu1261A | hydrogen bond donor |
| Arg880A | hydrogen bond donor, electrostatic stabiliser |
| Glu802A | hydrogen bond acceptor, electrostatic stabiliser |
| Glu1261A | proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, electron transfer, radical termination, overall reactant used, cofactor used, native state of cofactor regenerated, coordination to a metal ion, decoordination from a metal ion, intermediate terminated, intermediate formation, overall product formed, electron relay
Step 4. FAD is regenerated through a hydroxide transfer from the FAD cofactor to the substrate NAD
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu1261A | hydrogen bond acceptor |
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, hydride transfer, ingold: aromatic bimolecular nucleophilic addition, overall reactant used, intermediate terminated, overall product formed, native state of enzyme regenerated, native state of cofactor regeneratedIntroduction
An alternative mechanism has been proposed for the formation of the first intermediate in which the mechanism of the reaction proceeds via individual one-electron steps (rather than the obligatory two-electron chemistry of a nucleophilic attack mechanism). However, the lack of inverse relationship between the one electron reduction potential for the purine substrates and the rate of catalysis suggests that the mechanism proceeds via the nucleophilic attack shown in the other proposal.
Catalytic Residues Roles
| UniProt | PDB* (1v97) | ||
| Glu802 | Glu802A | The residue is thought to influence the tautomerisation step, relaying a proton from Glu1261 to a nitrogen on the substrate by lowering the energy of the desired tautomer, with concomitant hydride transfer to the metal cofactor. | hydrogen bond acceptor, electrostatic stabiliser |
| Arg880 | Arg880A | The positively charged gaunidinium side chain acts to stabilise the build up of negative charge on the substrate once nucleophilic attack has occurred. | hydrogen bond donor, electrostatic stabiliser |
| Glu1261 | Glu1261A | Thought to stabilise the reactive intermediates and transition states formed during the course of the reaction in this proposal. | hydrogen bond acceptor, electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic addition, overall reactant used, cofactor used, intermediate formation, proton transfer, intramolecular nucleophilic addition, electron transfer, hydride transfer, overall product formed, inferred reaction stepReferences
- Stockert AL et al. (2002), J Am Chem Soc, 124, 14554-14555. The Reaction Mechanism of Xanthine Oxidase: Evidence for Two-Electron Chemistry Rather Than Sequential One-Electron Steps. DOI:10.1021/ja027388d.
- Howes BD et al. (1996), Biochemistry, 35, 1432-1443. Evidence favoring molybdenum-carbon bond formation in xanthine oxidase action: 17Q- and 13C-ENDOR and kinetic studies. DOI:10.1021/bi9520500. PMID:8634273.
Step 1. The sulfur of MTE abstracts a proton from the substrate, which initiates a nucleophilic attack on the Mo(VI) to form Mo(IV) and a Mo-C bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu1261A | hydrogen bond acceptor, electrostatic stabiliser |
| Arg880A | hydrogen bond donor, electrostatic stabiliser |
| Glu802A | hydrogen bond acceptor, electrostatic stabiliser |
Chemical Components
ingold: bimolecular nucleophilic addition, overall reactant used, cofactor used, intermediate formation, proton transfer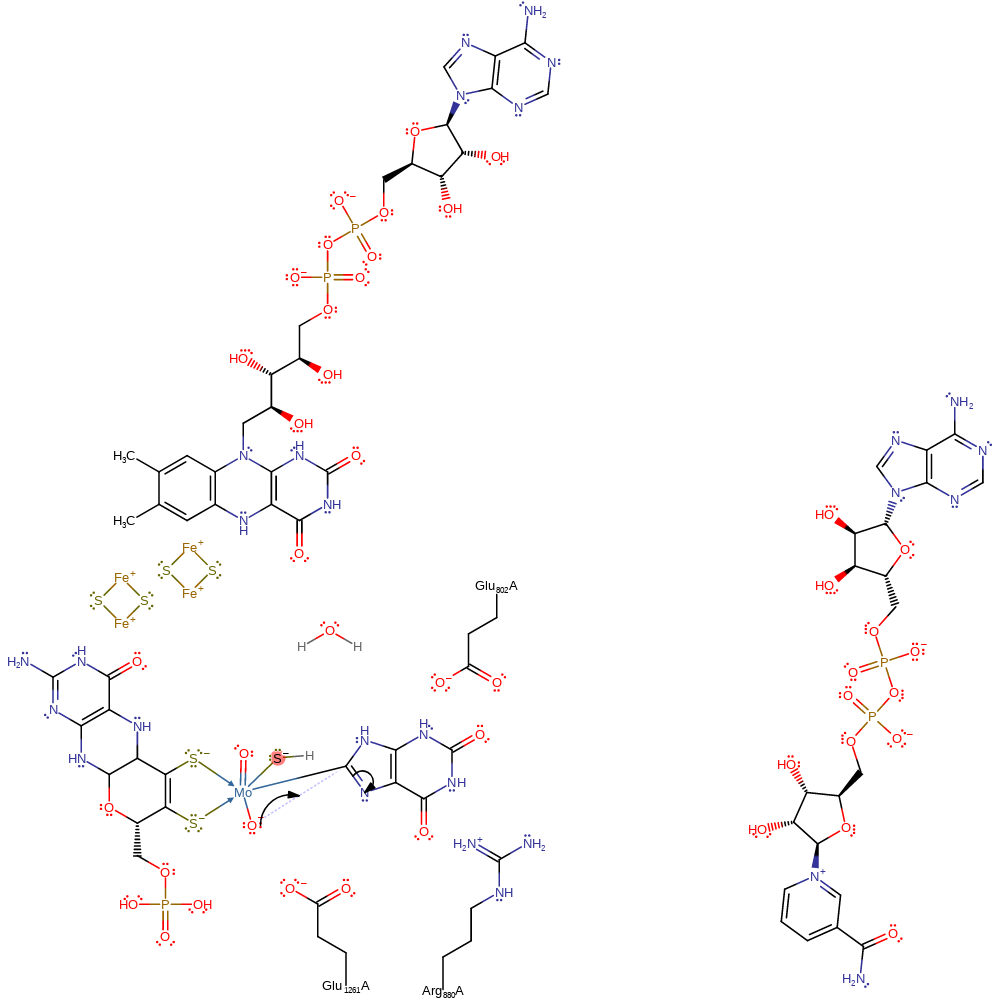
Step 2. The hydroxide ligand of the Mo cofactor attacks the carbon of the Mo-C bond, forming a three centre, two electon bond between Mo-C-O and a negatively charged nitrogen which is stabilised by the argenine residue.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu802A | electrostatic stabiliser |
| Arg880A | electrostatic stabiliser |
| Glu1261A | electrostatic stabiliser |
Chemical Components
ingold: intramolecular nucleophilic addition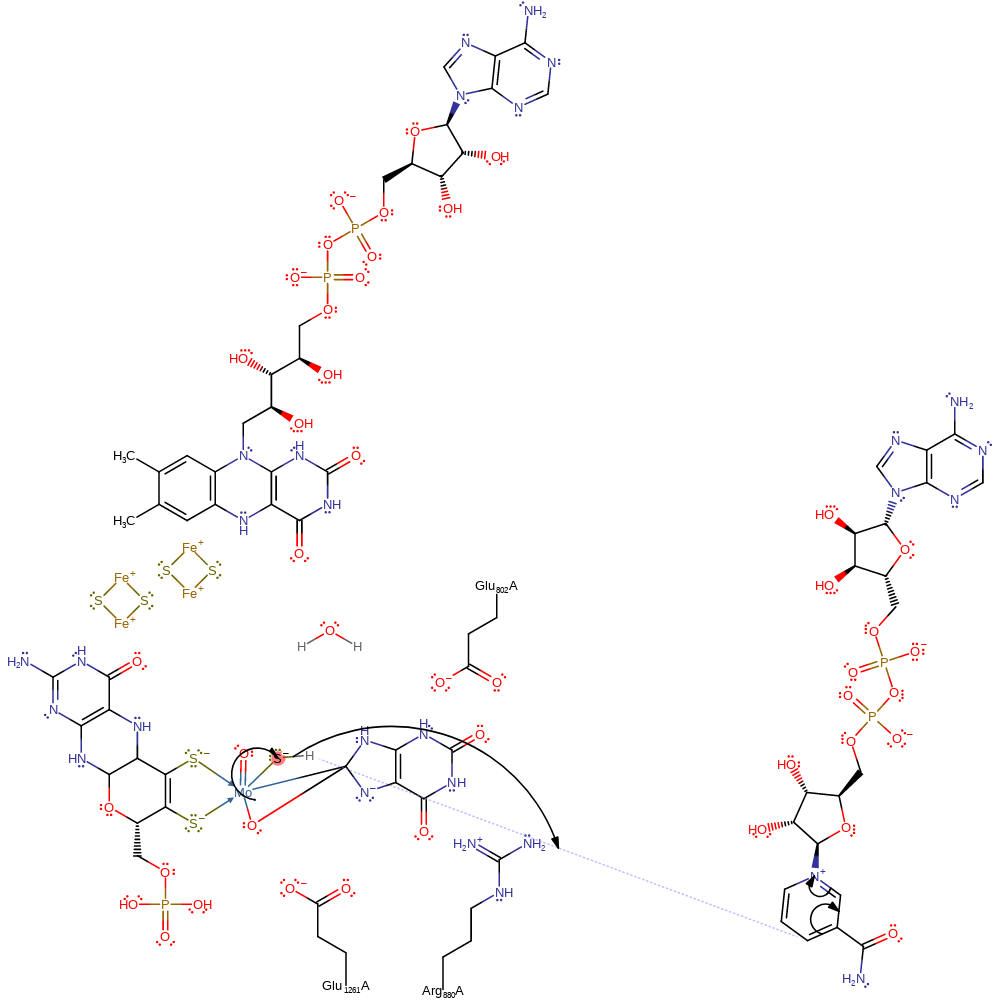
Step 3. A single proton and electron are lost from the Mo-thiol group and substrate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu802A | electrostatic stabiliser |
| Arg880A | electrostatic stabiliser |
| Glu1261A | electrostatic stabiliser |
Chemical Components
electron transfer, hydride transfer, overall product formed, overall reactant used, inferred reaction step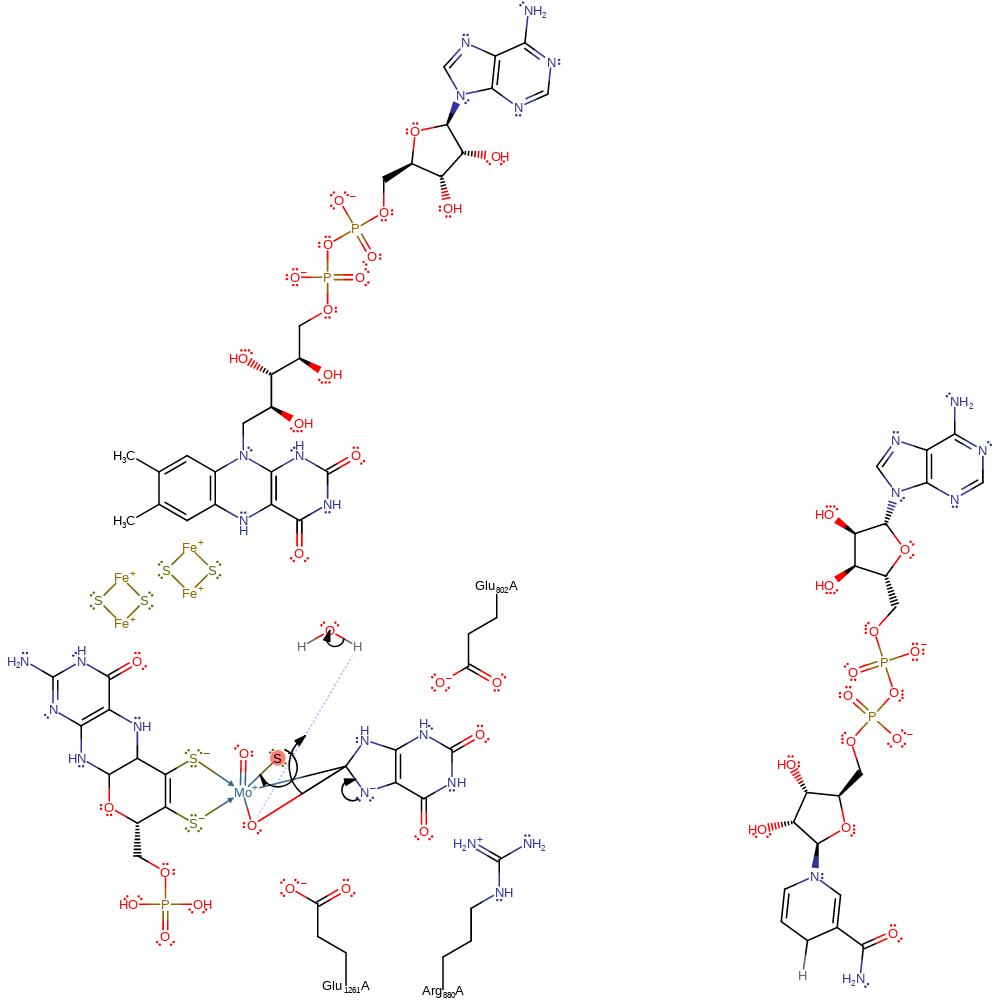
Step 4. The negatively charged nitrogen initiates a proton transfer from the active site water to the oxygen atom now bonded to the C8 of the intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu802A | electrostatic stabiliser |
| Arg880A | electrostatic stabiliser |
| Glu1261A | electrostatic stabiliser |
Chemical Components
overall product formed, inferred reaction step, electron transfer, proton transferIntroduction
A third proposal suggests that the first intermediate is formed via the addition of the C8-H bond across the Mo=O group of the molybdenum centre. However, the Mo-O-C bond in the product of this step has been crystallographically proven which rules out this alternative.
Catalytic Residues Roles
| UniProt | PDB* (1v97) | ||
| Glu802 | Glu802A | The residue is thought to influence the tautomerisation step. | hydrogen bond acceptor, electrostatic stabiliser |
| Arg880 | Arg880A | The positively charged gaunidinium side chain acts to stabilise the build up of negative charge on the substrate once nucleophilic attack has occurred. | hydrogen bond donor, electrostatic stabiliser |
| Glu1261 | Glu1261A | Helps stabilise the reactive intermediates and transition states formed during the course of the reaction. | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser, proton donor |
Chemical Components
proton transfer, bimolecular nucleophilic substitution, hydride transfer, bimolecular nucleophilic addition, overall reactant used, cofactor used, intermediate formation, redox reaction, radical formation, native state of cofactor regenerated, overall product formed, electron relay, electron transfer, radical termination, coordination to a metal ion, decoordination from a metal ion, intermediate terminated, aromatic unimolecular elimination by the conjugate base, aromatic bimolecular nucleophilic addition, native state of enzyme regeneratedReferences
- Xia M et al. (1999), J Biol Chem, 274, 3323-3330. The Reductive Half-reaction of Xanthine Oxidase. DOI:10.1074/jbc.274.6.3323.
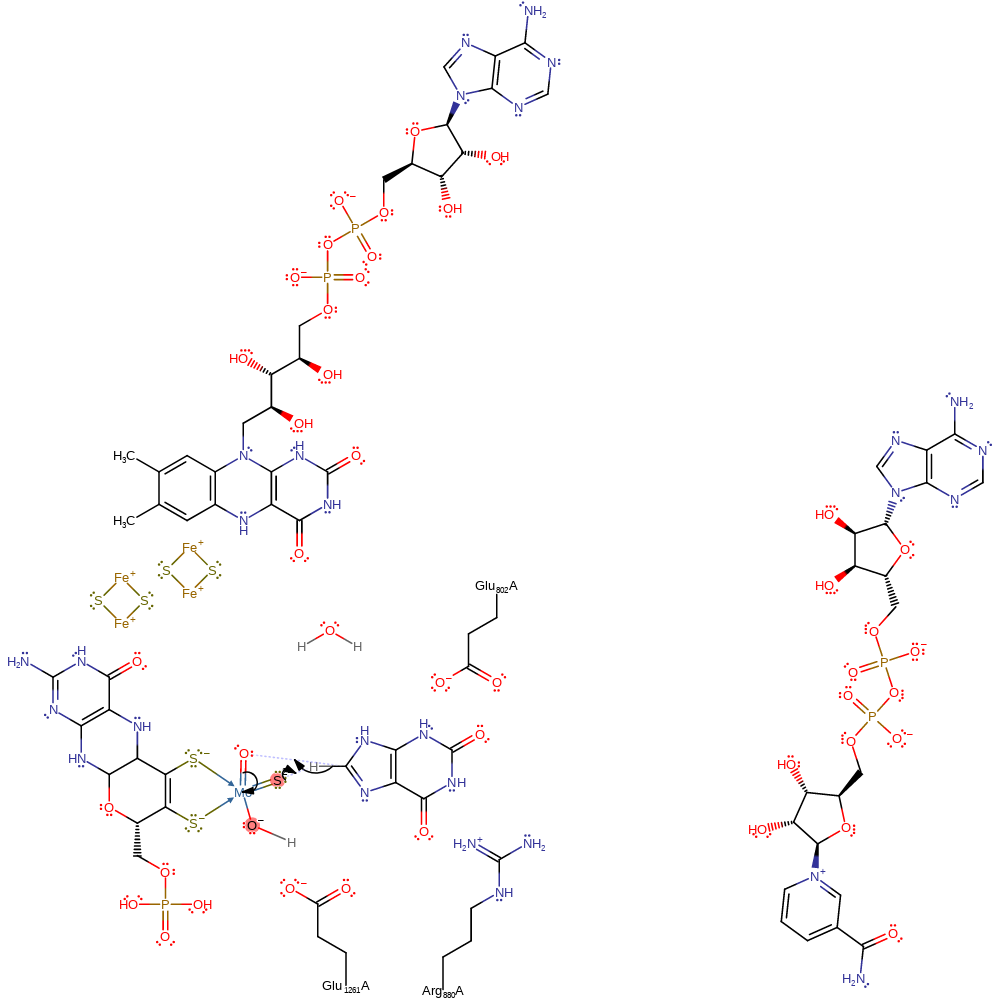
Step 1. The sulfur of the the MTE cofactor abstracts the proton from the C8 of the substrate. This initiates a nucleophilic attack upon the Mo=O ligand, resulting in a two electron reduction of the Mo(VI) to Mo(IV) and a thiol bound to molybdenum.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu1261A | hydrogen bond acceptor, electrostatic stabiliser |
| Arg880A | hydrogen bond donor, electrostatic stabiliser |
| Glu802A | hydrogen bond acceptor, electrostatic stabiliser |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, hydride transfer, ingold: bimolecular nucleophilic addition, overall reactant used, cofactor used, intermediate formation
Step 2. Water deprotonates the molybdenum bound thiol, reforming the Mo=S species and a single electron transfer from the Mo(IV) through the rest of the cofactor and two iron-sulfur clusters to a bound FAD cofactor, forming Mo(V) and a radical on the FAD cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu1261A | hydrogen bond donor, electrostatic stabiliser |
| Arg880A | hydrogen bond donor |
| Glu802A | hydrogen bond acceptor |
Chemical Components
proton transfer, redox reaction, radical formation, cofactor used, native state of cofactor regenerated, intermediate formation, overall product formed, electron relay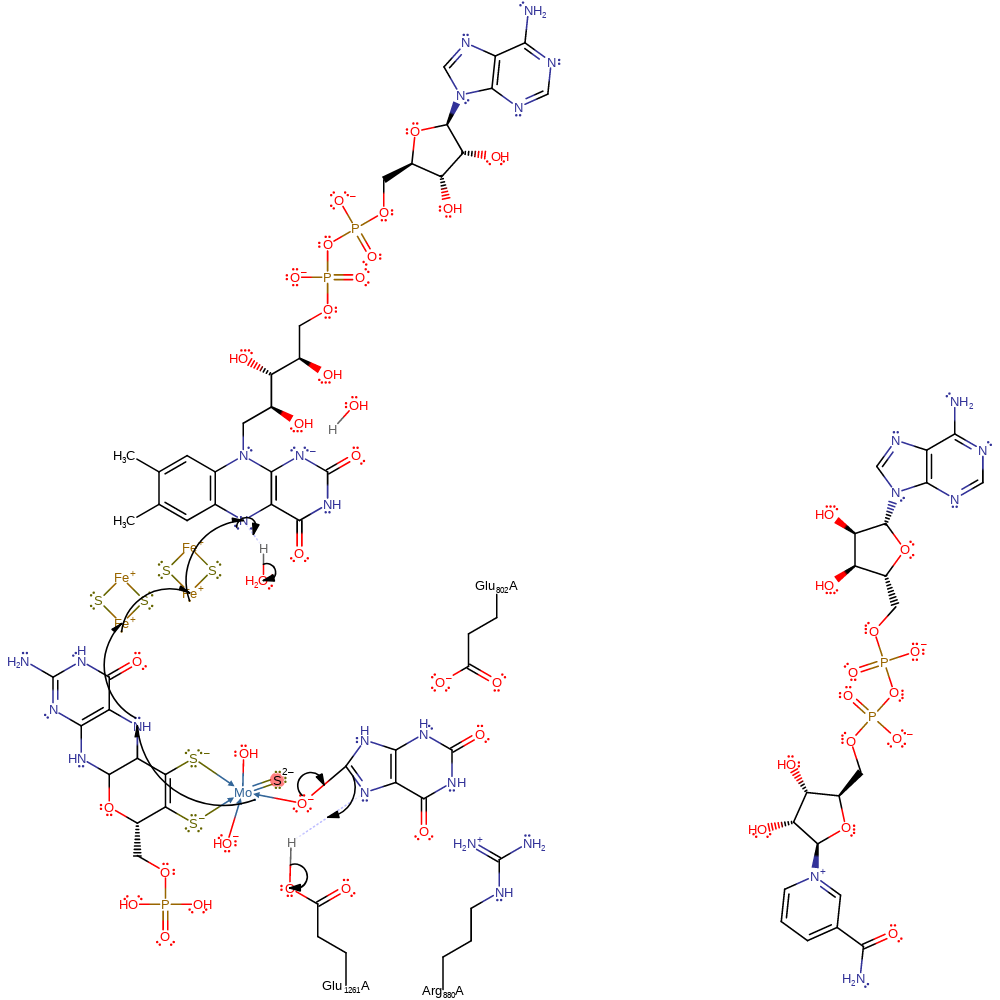
Step 3. A free hydroxide ion attacks the Mo(V), releasing the urate product (oxidised xanthine), which is re-protonated from the Glu1261, in a nucleophilic substitution reaction. This also results in the second electron transfer from the Mo(V) through the rest of the cofactor and two iron-sulfur clusters to a bound FAD cofactor, which then deprotonates a hydroxonium ion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu1261A | hydrogen bond donor |
| Arg880A | hydrogen bond donor, electrostatic stabiliser |
| Glu802A | hydrogen bond acceptor, electrostatic stabiliser |
| Glu1261A | proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, electron transfer, radical termination, overall reactant used, cofactor used, native state of cofactor regenerated, coordination to a metal ion, decoordination from a metal ion, intermediate terminated, intermediate formation, overall product formed, electron relay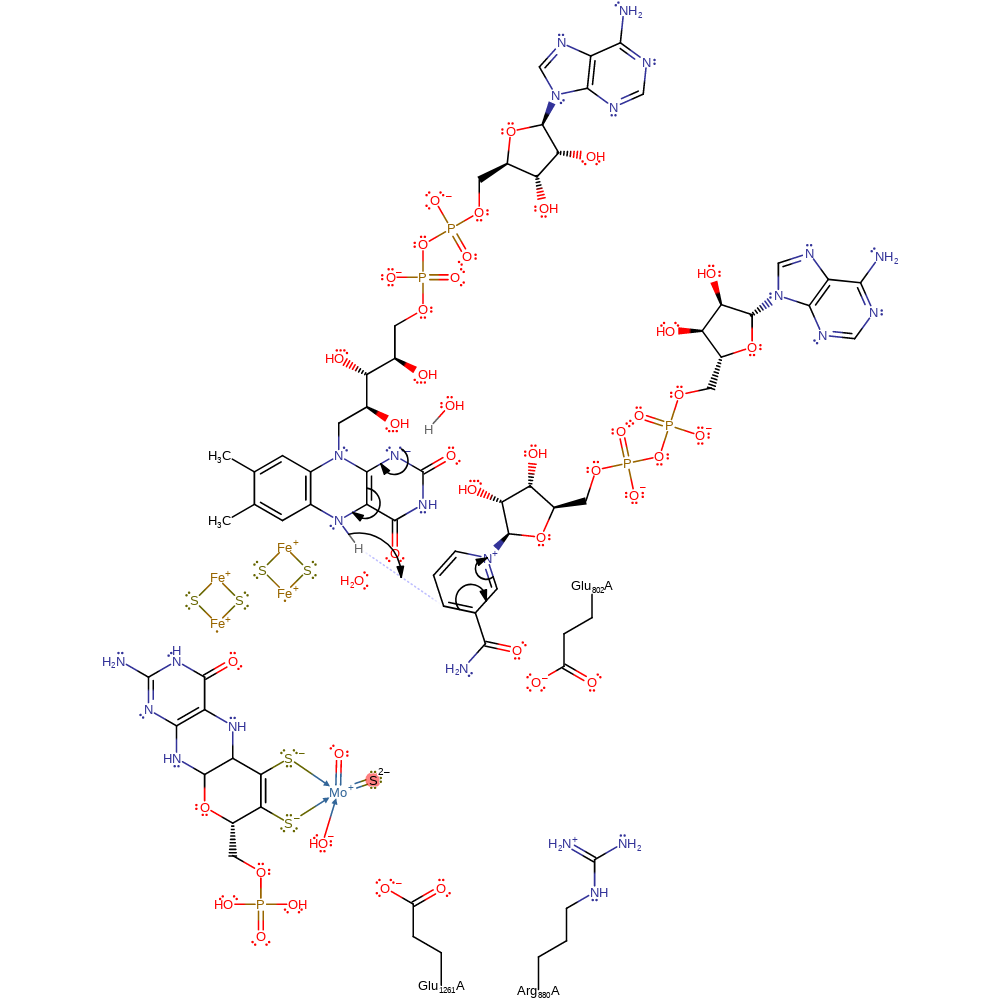
Step 4. FAD is regenerated through a hydroxide transfer from the FAD cofactor to the substrate NAD
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu1261A | hydrogen bond acceptor |






 Download:
Download: 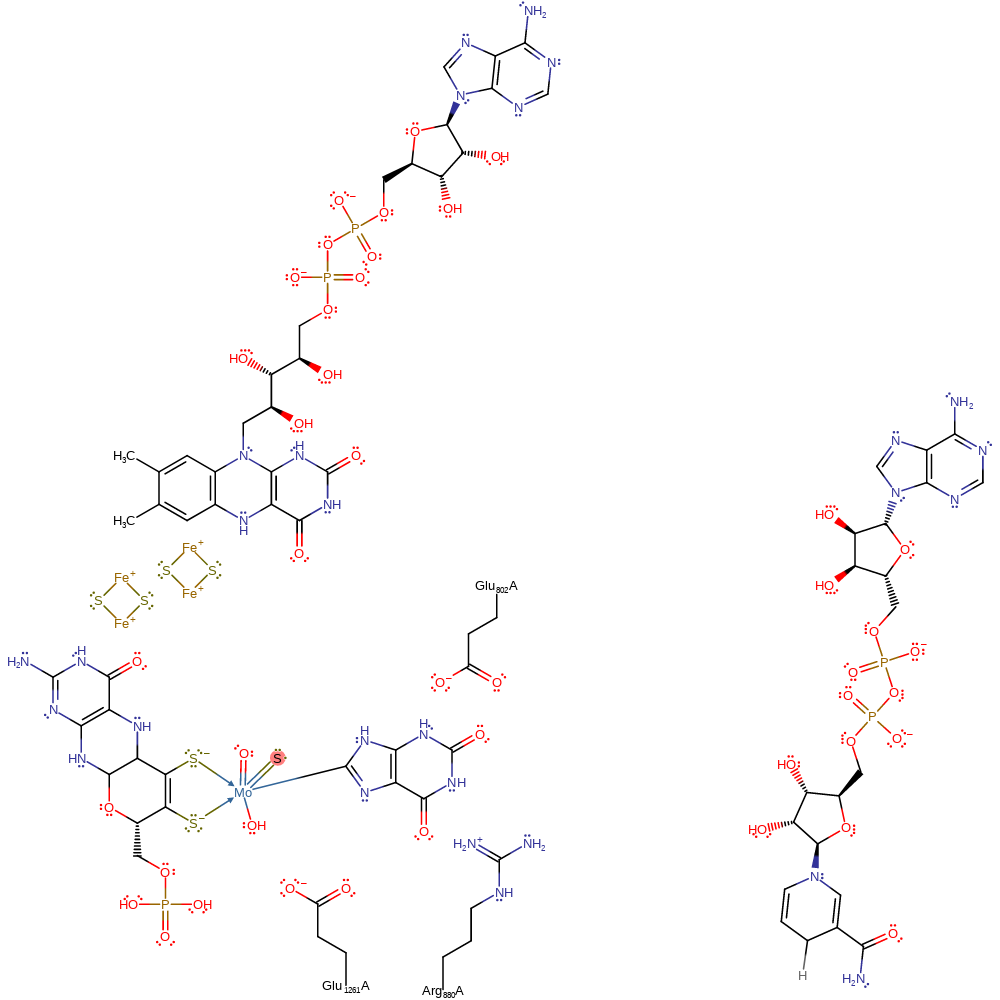 Download:
Download: 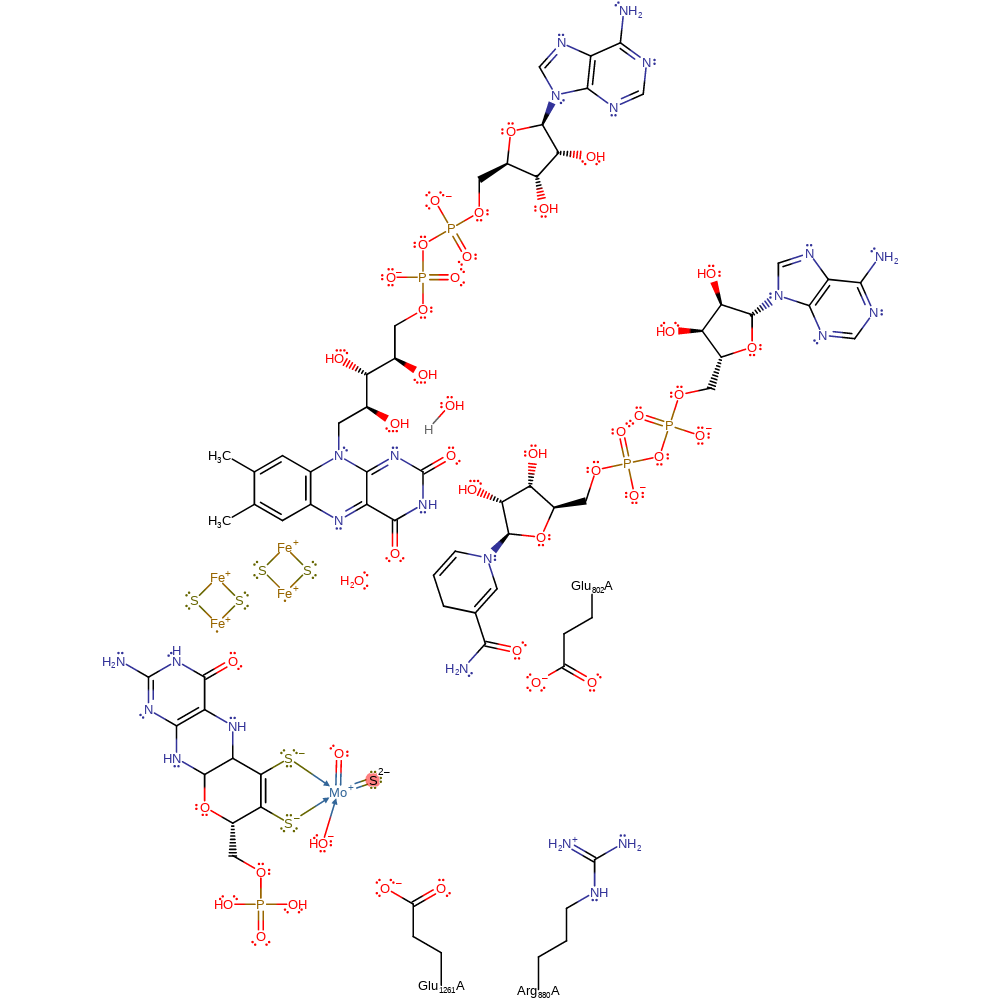 Download:
Download: