 |
PDBsum entry 1v97

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1v97
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.17.1.4
- xanthine dehydrogenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Xanthine Dehydrogenase
|
 |
 |
 |
 |
 |
Reaction:
|
 |
xanthine + NAD+ + H2O = urate + NADH + H+
|
 |
 |
 |
 |
 |
xanthine
Bound ligand (Het Group name = )
matches with 45.83% similarity
|
+
|
 NAD(+)
NAD(+)
|
+
|
H2O
|
=
|
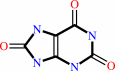 urate
urate
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD; Iron-sulfur; Mo cation
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
 Iron-sulfur
Iron-sulfur
|
Mo cation
|
|
 |
 |
Enzyme class 2:
|
 |
E.C.1.17.3.2
- xanthine oxidase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
xanthine + O2 + H2O = urate + H2O2
|
 |
 |
 |
 |
 |
xanthine
Bound ligand (Het Group name = )
matches with 45.83% similarity
|
+
|
 O2
O2
|
+
|
H2O
|
=
|
 urate
urate
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD; Iron-sulfur; Mo-molybdopterin
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
 Iron-sulfur
Iron-sulfur
|
Mo-molybdopterin
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
101:7931-7936
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
The crystal structure of xanthine oxidoreductase during catalysis: implications for reaction mechanism and enzyme inhibition.
|
|
K.Okamoto,
K.Matsumoto,
R.Hille,
B.T.Eger,
E.F.Pai,
T.Nishino.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Molybdenum is widely distributed in biology and is usually found as a
mononuclear metal center in the active sites of many enzymes catalyzing oxygen
atom transfer. The molybdenum hydroxylases are distinct from other biological
systems catalyzing hydroxylation reactions in that the oxygen atom incorporated
into the product is derived from water rather than molecular oxygen. Here, we
present the crystal structure of the key intermediate in the hydroxylation
reaction of xanthine oxidoreductase with a slow substrate, in which the
carbon-oxygen bond of the product is formed, yet the product remains complexed
to the molybdenum. This intermediate displays a stable broad charge-transfer
band at approximately 640 nm. The crystal structure of the complex indicates
that the catalytically labile Mo-OH oxygen has formed a bond with a carbon atom
of the substrate. In addition, the MoS group of the oxidized enzyme has become
protonated to afford Mo-SH on reduction of the molybdenum center. In contrast to
previous assignments, we find this last ligand at an equatorial position in the
square-pyramidal metal coordination sphere, not the apical position. A water
molecule usually seen in the active site of the enzyme is absent in the present
structure, which probably accounts for the stability of this intermediate toward
ligand displacement by hydroxide.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Fig. 4. Stereo representation of the structure in the
active site of XDH with bound FYX-051. FYX-051 (magenta),
molybdopterin (cyan), and catalytically important amino acid
residues (CPK-atom colored) are illustrated as stick models on a
ribbon model background.
|
 |
Figure 7.
Fig. 7. Proposed mechanism initiated by base-assisted
nucleophilic attack of Mo--OH on heterocycles, with subsequent
hydride transfer to produce the reaction intermediate (c) whose
structure has been analyzed in this report. The subsequent
oxidation occurs via d or/and e with varying ratio depending on
the substrate used.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Jin,
A.J.Straathof,
M.W.Pinkse,
and
U.Hanefeld
(2011).
Purification, characterization, and cloning of a bifunctional molybdoenzyme with hydratase and alcohol dehydrogenase activity.
|
| |
Appl Microbiol Biotechnol,
89,
1831-1840.
|
 |
|
|
|
|
 |
L.B.Maia,
and
J.J.Moura
(2011).
Nitrite reduction by xanthine oxidase family enzymes: a new class of nitrite reductases.
|
| |
J Biol Inorg Chem,
16,
443-460.
|
 |
|
|
|
|
 |
M.Leigh,
C.E.Castillo,
D.J.Raines,
and
A.K.Duhme-Klair
(2011).
Synthesis, activity testing and molybdenum(VI) complexation of Schiff bases derived from 2,4,6-trihydroxybenzaldehyde investigated as xanthine oxidase inhibitors.
|
| |
ChemMedChem,
6,
612-616.
|
 |
|
|
|
|
 |
N.Ashizawa,
T.Shimo,
K.Matsumoto,
T.Taniguchi,
M.Moto,
and
O.Nagata
(2011).
Establishment of simultaneous treatment model with citrate for preventing nephropathy induced by FYX-051, a xanthine oxidoreductase inhibitor, in rats.
|
| |
Drug Chem Toxicol,
34,
151-161.
|
 |
|
|
|
|
 |
T.Shimo,
N.Ashizawa,
M.Moto,
T.Iwanaga,
and
O.Nagata
(2011).
Study on species differences in nephropathy induced by FYX-051, a xanthine oxidoreductase inhibitor.
|
| |
Arch Toxicol,
85,
505-512.
|
 |
|
|
|
|
 |
L.M.Blank,
B.E.Ebert,
K.Buehler,
and
B.Bühler
(2010).
Redox biocatalysis and metabolism: molecular mechanisms and metabolic network analysis.
|
| |
Antioxid Redox Signal,
13,
349-394.
|
 |
|
|
|
|
 |
J.F.Alfaro,
C.A.Joswig-Jones,
W.Ouyang,
J.Nichols,
G.J.Crouch,
and
J.P.Jones
(2009).
Purification and mechanism of human aldehyde oxidase expressed in Escherichia coli.
|
| |
Drug Metab Dispos,
37,
2393-2398.
|
 |
|
|
|
|
 |
M.J.Romão
(2009).
Molybdenum and tungsten enzymes: a crystallographic and mechanistic overview.
|
| |
Dalton Trans,
(),
4053-4068.
|
 |
|
|
|
|
 |
U.Dietzel,
J.Kuper,
J.A.Doebbler,
A.Schulte,
J.J.Truglio,
S.Leimkühler,
and
C.Kisker
(2009).
Mechanism of Substrate and Inhibitor Binding of Rhodobacter capsulatus Xanthine Dehydrogenase.
|
| |
J Biol Chem,
284,
8768-8776.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.F.Alfaro,
and
J.P.Jones
(2008).
Studies on the mechanism of aldehyde oxidase and xanthine oxidase.
|
| |
J Org Chem,
73,
9469-9472.
|
 |
|
|
|
|
 |
K.Okamoto,
and
T.Nishino
(2008).
Crystal structures of mammalian xanthine oxidoreductase bound with various inhibitors: allopurinol, febuxostat, and FYX-051.
|
| |
J Nippon Med Sch,
75,
2-3.
|
 |
|
|
|
|
 |
M.Li,
T.A.Müller,
B.A.Fraser,
and
R.P.Hausinger
(2008).
Characterization of active site variants of xanthine hydroxylase from Aspergillus nidulans.
|
| |
Arch Biochem Biophys,
470,
44-53.
|
 |
|
|
|
|
 |
Q.Gao,
and
J.S.Thorson
(2008).
The biosynthetic genes encoding for the production of the dynemicin enediyne core in Micromonospora chersina ATCC53710.
|
| |
FEMS Microbiol Lett,
282,
105-114.
|
 |
|
|
|
|
 |
S.Chaves,
M.Gil,
S.Canário,
R.Jelic,
M.J.Romão,
J.Trincão,
E.Herdtweck,
J.Sousa,
C.Diniz,
P.Fresco,
and
M.A.Santos
(2008).
Biologically relevant O,S-donor compounds. Synthesis, molybdenum complexation and xanthine oxidase inhibition.
|
| |
Dalton Trans,
(),
1773-1782.
|
 |
|
|
|
|
 |
T.Nishino,
K.Okamoto,
B.T.Eger,
E.F.Pai,
and
T.Nishino
(2008).
Mammalian xanthine oxidoreductase - mechanism of transition from xanthine dehydrogenase to xanthine oxidase.
|
| |
FEBS J,
275,
3278-3289.
|
 |
|
|
|
|
 |
A.Thapper,
D.R.Boer,
C.D.Brondino,
J.J.Moura,
and
M.J.Romão
(2007).
Correlating EPR and X-ray structural analysis of arsenite-inhibited forms of aldehyde oxidoreductase.
|
| |
J Biol Inorg Chem,
12,
353-366.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.M.Montero-Morán,
M.Li,
E.Rendòn-Huerta,
F.Jourdan,
D.J.Lowe,
A.W.Stumpff-Kane,
M.Feig,
C.Scazzocchio,
and
R.P.Hausinger
(2007).
Purification and characterization of the FeII- and alpha-ketoglutarate-dependent xanthine hydroxylase from Aspergillus nidulans.
|
| |
Biochemistry,
46,
5293-5304.
|
 |
|
|
|
|
 |
S.Kalra,
G.Jena,
K.Tikoo,
and
A.K.Mukhopadhyay
(2007).
Preferential inhibition of xanthine oxidase by 2-amino-6-hydroxy-8-mercaptopurine and 2-amino-6-purine thiol.
|
| |
BMC Biochem,
8,
8.
|
 |
|
|
|
|
 |
Y.C.Chang,
F.W.Lee,
C.S.Chen,
S.T.Huang,
S.H.Tsai,
S.H.Huang,
and
C.M.Lin
(2007).
Structure-activity relationship of C6-C3 phenylpropanoids on xanthine oxidase-inhibiting and free radical-scavenging activities.
|
| |
Free Radic Biol Med,
43,
1541-1551.
|
 |
|
|
|
|
 |
C.D.Brondino,
M.J.Romão,
I.Moura,
and
J.J.Moura
(2006).
Molybdenum and tungsten enzymes: the xanthine oxidase family.
|
| |
Curr Opin Chem Biol,
10,
109-114.
|
 |
|
|
|
|
 |
G.Schwarz,
and
R.R.Mendel
(2006).
Molybdenum cofactor biosynthesis and molybdenum enzymes.
|
| |
Annu Rev Plant Biol,
57,
623-647.
|
 |
|
|
|
|
 |
H.Tamta,
S.Kalra,
and
A.K.Mukhopadhyay
(2006).
Biochemical characterization of some pyrazolopyrimidine-based inhibitors of xanthine oxidase.
|
| |
Biochemistry (Mosc),
71,
S49-S54.
|
 |
|
|
|
|
 |
N.Ashizawa,
T.Shimo,
K.Matsumoto,
K.Oba,
T.Nakazawa,
and
O.Nagata
(2006).
Strain differences in the responsiveness between Sprague-Dawley and Fischer rats to nephropathy induced by FYX-051, a xanthine oxidoreductase inhibitor.
|
| |
Toxicol Appl Pharmacol,
217,
260-265.
|
 |
|
|
|
|
 |
P.Pacher,
A.Nivorozhkin,
and
C.Szabó
(2006).
Therapeutic effects of xanthine oxidase inhibitors: renaissance half a century after the discovery of allopurinol.
|
| |
Pharmacol Rev,
58,
87.
|
 |
|
|
|
|
 |
A.Cultrone,
C.Scazzocchio,
M.Rochet,
G.Montero-Morán,
C.Drevet,
and
R.Fernández-Martín
(2005).
Convergent evolution of hydroxylation mechanisms in the fungal kingdom: molybdenum cofactor-independent hydroxylation of xanthine via alpha-ketoglutarate-dependent dioxygenases.
|
| |
Mol Microbiol,
57,
276-290.
|
 |
|
|
|
|
 |
D.R.Boer,
A.Müller,
S.Fetzner,
D.J.Lowe,
and
M.J.Romão
(2005).
On the purification and preliminary crystallographic analysis of isoquinoline 1-oxidoreductase from Brevundimonas diminuta 7.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
137-140.
|
 |
|
|
|
|
 |
H.Tamta,
R.Thilagavathi,
A.K.Chakraborti,
and
A.K.Mukhopadhyay
(2005).
6-(N-benzoylamino)purine as a novel and potent inhibitor of xanthine oxidase: inhibition mechanism and molecular modeling studies.
|
| |
J Enzyme Inhib Med Chem,
20,
317-324.
|
 |
|
|
|
|
 |
M.Resch,
H.Dobbek,
and
O.Meyer
(2005).
Structural and functional reconstruction in situ of the [CuSMoO2] active site of carbon monoxide dehydrogenase from the carbon monoxide oxidizing eubacterium Oligotropha carboxidovorans.
|
| |
J Biol Inorg Chem,
10,
518-528.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

