 |
PDBsum entry 1daa

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase (aminotransferase)
|
PDB id
|
|

|

|
1daa
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.6.1.21
- D-amino-acid transaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-alanine + 2-oxoglutarate = D-glutamate + pyruvate
|
 |
 |
 |
 |
 |
 D-alanine
D-alanine
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
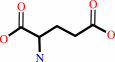 D-glutamate
D-glutamate
|
+
|
 pyruvate
pyruvate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
34:9661-9669
(1995)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of a D-amino acid aminotransferase: how the protein controls stereoselectivity.
|
|
S.Sugio,
G.A.Petsko,
J.M.Manning,
K.Soda,
D.Ringe.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The three-dimensional structure of D-amino acid aminotransferase (D-AAT) in the
pyridoxamine phosphate form has been determined crystallographically. The fold
of this pyridoxal phosphate (PLP)-containing enzyme is completely different from
those of any of the other enzymes that utilize PLP as part of their mechanism
and whose structures are known. However, there are some striking similarities
between the active sites of D-AAT and the corresponding enzyme that
transaminates L-amino acids, L-aspartate aminotransferase. These similarities
represent convergent evolution to a common solution of the problem of enforcing
transamination chemistry on the PLP cofactor. Implications of these similarities
are discussed in terms of their possible roles in the stabilization of
intermediates of a transamination reaction. In addition, sequence similarity
between D-AAT and branched chain L-amino acid aminotransferase suggests that
this latter enzyme will also have a fold similar to that of D-AAT.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Höhne,
S.Schätzle,
H.Jochens,
K.Robins,
and
U.T.Bornscheuer
(2010).
Rational assignment of key motifs for function guides in silico enzyme identification.
|
| |
Nat Chem Biol,
6,
807-813.
|
 |
|
|
|
|
 |
J.P.Richard,
T.L.Amyes,
J.Crugeiras,
and
A.Rios
(2009).
Pyridoxal 5'-phosphate: electrophilic catalyst extraordinaire.
|
| |
Curr Opin Chem Biol,
13,
475-483.
|
 |
|
|
|
|
 |
H.Barreteau,
A.Kovac,
A.Boniface,
M.Sova,
S.Gobec,
and
D.Blanot
(2008).
Cytoplasmic steps of peptidoglycan biosynthesis.
|
| |
FEMS Microbiol Rev,
32,
168-207.
|
 |
|
|
|
|
 |
M.Funakoshi,
M.Sekine,
M.Katane,
T.Furuchi,
M.Yohda,
T.Yoshikawa,
and
H.Homma
(2008).
Cloning and functional characterization of Arabidopsis thaliana D-amino acid aminotransferase--D-aspartate behavior during germination.
|
| |
FEBS J,
275,
1188-1200.
|
 |
|
|
|
|
 |
S.G.Lee,
S.P.Hong,
J.J.Song,
S.J.Kim,
M.S.Kwak,
and
M.H.Sung
(2006).
Functional and structural characterization of thermostable D-amino acid aminotransferases from Geobacillus spp.
|
| |
Appl Environ Microbiol,
72,
1588-1594.
|
 |
|
|
|
|
 |
G.W.Han,
R.Schwarzenbacher,
R.Page,
L.Jaroszewski,
P.Abdubek,
E.Ambing,
T.Biorac,
J.M.Canaves,
H.J.Chiu,
X.Dai,
A.M.Deacon,
M.DiDonato,
M.A.Elsliger,
A.Godzik,
C.Grittini,
S.K.Grzechnik,
J.Hale,
E.Hampton,
J.Haugen,
M.Hornsby,
H.E.Klock,
E.Koesema,
A.Kreusch,
P.Kuhn,
S.A.Lesley,
I.Levin,
D.McMullan,
T.M.McPhillips,
M.D.Miller,
A.Morse,
K.Moy,
E.Nigoghossian,
J.Ouyang,
J.Paulsen,
K.Quijano,
R.Reyes,
E.Sims,
G.Spraggon,
R.C.Stevens,
H.van den Bedem,
J.Velasquez,
J.Vincent,
F.von Delft,
X.Wang,
B.West,
A.White,
G.Wolf,
Q.Xu,
O.Zagnitko,
K.O.Hodgson,
J.Wooley,
and
I.A.Wilson
(2005).
Crystal structure of an alanine-glyoxylate aminotransferase from Anabaena sp. at 1.70 A resolution reveals a noncovalently linked PLP cofactor.
|
| |
Proteins,
58,
971-975.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Hirotsu,
M.Goto,
A.Okamoto,
and
I.Miyahara
(2005).
Dual substrate recognition of aminotransferases.
|
| |
Chem Rec,
5,
160-172.
|
 |
|
|
|
|
 |
M.Goto,
I.Miyahara,
K.Hirotsu,
M.Conway,
N.Yennawar,
M.M.Islam,
and
S.M.Hutson
(2005).
Structural determinants for branched-chain aminotransferase isozyme-specific inhibition by the anticonvulsant drug gabapentin.
|
| |
J Biol Chem,
280,
37246-37256.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.C.Eliot,
and
J.F.Kirsch
(2004).
Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations.
|
| |
Annu Rev Biochem,
73,
383-415.
|
 |
|
|
|
|
 |
P.Kongsaeree,
C.Samanchart,
P.Laowanapiban,
S.Wiyakrutta,
and
V.Meevootisom
(2003).
Crystallization and preliminary X-ray crystallographic analysis of d-phenylglycine aminotransferase from Pseudomonas stutzeri ST201.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
953-954.
|
 |
|
|
|
|
 |
H.Kagamiyama,
and
H.Hayashi
(2001).
Release of enzyme strain during catalysis reduces the activation energy barrier.
|
| |
Chem Rec,
1,
385-394.
|
 |
|
|
|
|
 |
K.Soda,
T.Yoshimura,
and
N.Esaki
(2001).
Stereospecificity for the hydrogen transfer of pyridoxal enzyme reactions.
|
| |
Chem Rec,
1,
373-384.
|
 |
|
|
|
|
 |
N.Yennawar,
J.Dunbar,
M.Conway,
S.Hutson,
and
G.Farber
(2001).
The structure of human mitochondrial branched-chain aminotransferase.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
506-515.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Christen,
and
P.K.Mehta
(2001).
From cofactor to enzymes. The molecular evolution of pyridoxal-5'-phosphate-dependent enzymes.
|
| |
Chem Rec,
1,
436-447.
|
 |
|
|
|
|
 |
A.Gutierrez,
T.Yoshimura,
Y.Fuchikami,
and
N.Esaki
(2000).
Modulation of activity and substrate specificity by modifying the backbone length of the distant interdomain loop of D-amino acid aminotransferase.
|
| |
Eur J Biochem,
267,
7218-7223.
|
 |
|
|
|
|
 |
D.Saadat,
and
D.H.Harrison
(2000).
Mirroring perfection: the structure of methylglyoxal synthase complexed with the competitive inhibitor 2-phosphoglycolate.
|
| |
Biochemistry,
39,
2950-2960.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
F.J.Ruzicka,
K.W.Lieder,
and
P.A.Frey
(2000).
Lysine 2,3-aminomutase from Clostridium subterminale SB4: mass spectral characterization of cyanogen bromide-treated peptides and cloning, sequencing, and expression of the gene kamA in Escherichia coli.
|
| |
J Bacteriol,
182,
469-476.
|
 |
|
|
|
|
 |
G.Schneider,
H.Käck,
and
Y.Lindqvist
(2000).
The manifold of vitamin B6 dependent enzymes.
|
| |
Structure,
8,
R1-R6.
|
 |
|
|
|
|
 |
K.Kishimoto,
C.Yasuda,
and
J.M.Manning
(2000).
Reversible dissociation/association of D-amino acid transaminase subunits: properties of isolated active dimers and inactive monomers.
|
| |
Biochemistry,
39,
381-387.
|
 |
|
|
|
|
 |
A.D.Kern,
M.A.Oliveira,
P.Coffino,
and
M.L.Hackert
(1999).
Structure of mammalian ornithine decarboxylase at 1.6 A resolution: stereochemical implications of PLP-dependent amino acid decarboxylases.
|
| |
Structure,
7,
567-581.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Watanabe,
Y.Kurokawa,
T.Yoshimura,
T.Kurihara,
K.Soda,
N.Esaki,
and
A.Watababe
(1999).
Role of lysine 39 of alanine racemase from Bacillus stearothermophilus that binds pyridoxal 5'-phosphate. Chemical rescue studies of Lys39 --> Ala mutant.
|
| |
J Biol Chem,
274,
4189-4194.
|
 |
|
|
|
|
 |
A.Yamashita,
H.Kato,
S.Wakatsuki,
T.Tomizaki,
T.Nakatsu,
K.Nakajima,
T.Hashimoto,
Y.Yamada,
and
J.Oda
(1999).
Structure of tropinone reductase-II complexed with NADP+ and pseudotropine at 1.9 A resolution: implication for stereospecific substrate binding and catalysis.
|
| |
Biochemistry,
38,
7630-7637.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.S.Ro,
and
E.W.Miles
(1999).
Structure and function of the tryptophan synthase alpha(2)beta(2) complex. Roles of beta subunit histidine 86.
|
| |
J Biol Chem,
274,
36439-36445.
|
 |
|
|
|
|
 |
K.A.Denessiouk,
A.I.Denesyuk,
J.V.Lehtonen,
T.Korpela,
and
M.S.Johnson
(1999).
Common structural elements in the architecture of the cofactor-binding domains in unrelated families of pyridoxal phosphate-dependent enzymes.
|
| |
Proteins,
35,
250-261.
|
 |
|
|
|
|
 |
M.Faure,
F.Glomot,
R.Bledsoe,
S.Hutson,
and
I.Papet
(1999).
Purification and cloning of the mitochondrial branched-chain amino acid aminotransferase from sheep placenta.
|
| |
Eur J Biochem,
259,
104-111.
|
 |
|
|
|
|
 |
P.W.van Ophem,
D.Peisach,
S.D.Erickson,
K.Soda,
D.Ringe,
and
J.M.Manning
(1999).
Effects of the E177K mutation in D-amino acid transaminase. Studies on an essential coenzyme anchoring group that contributes to stereochemical fidelity.
|
| |
Biochemistry,
38,
1323-1331.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Peisach,
D.M.Chipman,
P.W.Van Ophem,
J.M.Manning,
and
D.Ringe
(1998).
Crystallographic study of steps along the reaction pathway of D-amino acid aminotransferase.
|
| |
Biochemistry,
37,
4958-4967.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Hayashi,
H.Mizuguchi,
and
H.Kagamiyama
(1998).
The imine-pyridine torsion of the pyridoxal 5'-phosphate Schiff base of aspartate aminotransferase lowers its pKa in the unliganded enzyme and is crucial for the successive increase in the pKa during catalysis.
|
| |
Biochemistry,
37,
15076-15085.
|
 |
|
|
|
|
 |
I.G.Fotheringham,
S.A.Bledig,
and
P.P.Taylor
(1998).
Characterization of the genes encoding D-amino acid transaminase and glutamate racemase, two D-glutamate biosynthetic enzymes of Bacillus sphaericus ATCC 10208.
|
| |
J Bacteriol,
180,
4319-4323.
|
 |
|
|
|
|
 |
J.Davoodi,
P.M.Drown,
R.K.Bledsoe,
R.Wallin,
G.D.Reinhart,
and
S.M.Hutson
(1998).
Overexpression and characterization of the human mitochondrial and cytosolic branched-chain aminotransferases.
|
| |
J Biol Chem,
273,
4982-4989.
|
 |
|
|
|
|
 |
J.N.Jansonius
(1998).
Structure, evolution and action of vitamin B6-dependent enzymes.
|
| |
Curr Opin Struct Biol,
8,
759-769.
|
 |
|
|
|
|
 |
K.H.Jhee,
P.McPhie,
H.S.Ro,
and
E.W.Miles
(1998).
Tryptophan synthase mutations that alter cofactor chemistry lead to mechanism-based inactivation.
|
| |
Biochemistry,
37,
14591-14604.
|
 |
|
|
|
|
 |
K.Nakajima,
A.Yamashita,
H.Akama,
T.Nakatsu,
H.Kato,
T.Hashimoto,
J.Oda,
and
Y.Yamada
(1998).
Crystal structures of two tropinone reductases: different reaction stereospecificities in the same protein fold.
|
| |
Proc Natl Acad Sci U S A,
95,
4876-4881.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.P.Taylor,
D.P.Pantaleone,
R.F.Senkpeil,
and
I.G.Fotheringham
(1998).
Novel biosynthetic approaches to the production of unnatural amino acids using transaminases.
|
| |
Trends Biotechnol,
16,
412-418.
|
 |
|
|
|
|
 |
P.W.van Ophem,
S.D.Erickson,
A.Martinez del Pozo,
I.Haller,
B.T.Chait,
T.Yoshimura,
K.Soda,
D.Ringe,
G.Petsko,
and
J.M.Manning
(1998).
Substrate inhibition of D-amino acid transaminase and protection by salts and by reduced nicotinamide adenine dinucleotide: isolation and initial characterization of a pyridoxo intermediate related to inactivation.
|
| |
Biochemistry,
37,
2879-2888.
|
 |
|
|
|
|
 |
A.L.Osterman,
H.B.Brooks,
J.Rizo,
and
M.A.Phillips
(1997).
Role of Arg-277 in the binding of pyridoxal 5'-phosphate to Trypanosoma brucei ornithine decarboxylase.
|
| |
Biochemistry,
36,
4558-4567.
|
 |
|
|
|
|
 |
H.Hayashi,
and
H.Kagamiyama
(1997).
Transient-state kinetics of the reaction of aspartate aminotransferase with aspartate at low pH reveals dual routes in the enzyme-substrate association process.
|
| |
Biochemistry,
36,
13558-13569.
|
 |
|
|
|
|
 |
J.P.Shaw,
G.A.Petsko,
and
D.Ringe
(1997).
Determination of the structure of alanine racemase from Bacillus stearothermophilus at 1.9-A resolution.
|
| |
Biochemistry,
36,
1329-1342.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Kataoka,
M.Ikemi,
T.Morikawa,
T.Miyoshi,
K.Nishi,
M.Wada,
H.Yamada,
and
S.Shimizu
(1997).
Isolation and characterization of D-threonine aldolase, a pyridoxal-5'-phosphate-dependent enzyme from Arthrobacter sp. DK-38.
|
| |
Eur J Biochem,
248,
385-393.
|
 |
|
|
|
|
 |
A.Eden,
G.Simchen,
and
N.Benvenisty
(1996).
Two yeast homologs of ECA39, a target for c-Myc regulation, code for cytosolic and mitochondrial branched-chain amino acid aminotransferases.
|
| |
J Biol Chem,
271,
20242-20245.
|
 |
|
|
|
|
 |
A.Martinez del Pozo,
P.W.van Ophem,
D.Ringe,
G.Petsko,
K.Soda,
and
J.M.Manning
(1996).
Interaction of pyridoxal 5'-phosphate with tryptophan-139 at the subunit interface of dimeric D-amino acid transaminase.
|
| |
Biochemistry,
35,
2112-2116.
|
 |
|
|
|
|
 |
K.H.Jhee,
T.Yoshimura,
N.Esaki,
and
K.Soda
(1996).
Stereospecificity of thermostable ornithine 5-aminotransferase for the hydrogen transfer in the L- and D-ornithine transamination.
|
| |
Biochemistry,
35,
9792-9796.
|
 |
|
|
|
|
 |
V.S.Stoll,
M.S.Kimber,
and
E.F.Pai
(1996).
Insights into substrate binding by D-2-ketoacid dehydrogenases from the structure of Lactobacillus pentosus D-lactate dehydrogenase.
|
| |
Structure,
4,
437-447.
|
 |
|
|
|
|
 |
W.M.Jones,
P.W.van Ophem,
M.A.Pospischil,
D.Ringe,
G.Petsko,
K.Soda,
and
J.M.Manning
(1996).
The ubiquitous cofactor NADH protects against substrate-induced inhibition of a pyridoxal enzyme.
|
| |
Protein Sci,
5,
2545-2551.
|
 |
|
|
|
|
 |
P.W.Van Ophem,
M.A.Pospischil,
D.Ringe,
D.Peisach,
G.Petsko,
K.Soda,
and
J.M.Manning
(1995).
Catalytic ability and stability of two recombinant mutants of D-amino acid transaminase involved in coenzyme binding.
|
| |
Protein Sci,
4,
2578-2586.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

