 |
PDBsum entry 2a1h

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
X-ray crystal structure of human mitochondrial branched chain aminotransferase (bcatm) complexed with gabapentin
|
|
Structure:
|
 |
Branched chain aminotransferase. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: mitochondrial bcat. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
1.80Å
|
R-factor:
|
0.208
|
R-free:
|
0.232
|
|
|
Authors:
|
 |
M.Goto,I.Miyahara,K.Hirotsu,M.Conway,N.Yennawar,M.M.Islam,S.M.Hutson
|
Key ref:

|
 |
M.Goto
et al.
(2005).
Structural determinants for branched-chain aminotransferase isozyme-specific inhibition by the anticonvulsant drug gabapentin.
J Biol Chem,
280,
37246-37256.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
20-Jun-05
|
Release date:
|
06-Sep-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O15382
(BCAT2_HUMAN) -
Branched-chain-amino-acid aminotransferase, mitochondrial from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
392 a.a.
363 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.6.1.42
- branched-chain-amino-acid transaminase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Leucine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-leucine + 2-oxoglutarate = 4-methyl-2-oxopentanoate + L-glutamate
|
 |
 |
 |
 |
 |
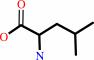 L-leucine
L-leucine
|
+
|
2-oxoglutarate
Bound ligand (Het Group name = )
matches with 46.67% similarity
|
=
|
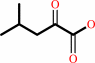 4-methyl-2-oxopentanoate
4-methyl-2-oxopentanoate
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
280:37246-37256
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural determinants for branched-chain aminotransferase isozyme-specific inhibition by the anticonvulsant drug gabapentin.
|
|
M.Goto,
I.Miyahara,
K.Hirotsu,
M.Conway,
N.Yennawar,
M.M.Islam,
S.M.Hutson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
This study presents the first three-dimensional structures of human cytosolic
branched-chain aminotransferase (hBCATc) isozyme complexed with the neuroactive
drug gabapentin, the hBCATc Michaelis complex with the substrate analog,
4-methylvalerate, and the mitochondrial isozyme (hBCATm) complexed with
gabapentin. The branched-chain aminotransferases (BCAT) reversibly catalyze
transamination of the essential branched-chain amino acids (leucine, isoleucine,
valine) to alpha-ketoglutarate to form the respective branched-chain alpha-keto
acids and glutamate. The cytosolic isozyme is the predominant BCAT found in the
nervous system, and only hBCATc is inhibited by gabapentin. Pre-steady state
kinetics show that 1.3 mm gabapentin can completely inhibit the binding of
leucine to reduced hBCATc, whereas 65.4 mm gabapentin is required to inhibit
leucine binding to hBCATm. Structural analysis shows that the bulky gabapentin
is enclosed in the active-site cavity by the shift of a flexible loop that
enlarges the active-site cavity. The specificity of gabapentin for the cytosolic
isozyme is ascribed at least in part to the location of the interdomain loop and
the relative orientation between the small and large domain which is different
from these relationships in the mitochondrial isozyme. Both isozymes contain a
CXXC center and form a disulfide bond under oxidizing conditions. The structure
of reduced hBCATc was obtained by soaking the oxidized hBCATc crystals with
dithiothreitol. The close similarity in active-site structures between cytosolic
enzyme complexes in the oxidized and reduced states is consistent with the small
effect of oxidation on pre-steady state kinetics of the hBCATc first
half-reaction. However, these kinetic data do not explain the inactivation of
hBCATm by oxidation of the CXXC center. The structural data suggest that there
is a larger effect of oxidation on the interdomain loop and residues surrounding
the CXXC center in hBCATm than in hBCATc.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
FIGURE 4. Schematic diagram showing hydrogen-bond and
salt-bridge interactions of the active-site residues. Putative
interactions are shown by dotted lines if the acceptor and donor
are less than 3.5 Å apart. The hydrogen bonds associated
with the phosphate group of PLP are omitted for clarity. A,
hBCATc-ox complexed with gabapentin. B, hBCATc-ox complexed with
4MeVA.
|
 |
Figure 6.
FIGURE 6. Stereo diagram of the superimposed active sites
of hBCATc-ox·gabapentin and hBCATc·gabapentin. The
residues and gabapentin, which represent
hBCATc-ox·gabapentin and hBCATc·gabapentin, are
shown in brown and deep blue, respectively.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2005,
280,
37246-37256)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Castell,
C.Mille,
and
T.Unge
(2010).
Structural analysis of mycobacterial branched-chain aminotransferase: implications for inhibitor design.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
549-557.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.W.Tremblay,
and
J.S.Blanchard
(2009).
The 1.9 A structure of the branched-chain amino-acid transaminase (IlvE) from Mycobacterium tuberculosis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
1071-1077.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.D.Chen,
T.F.Huang,
C.H.Lin,
H.H.Guan,
Y.C.Hsieh,
Y.H.Lin,
Y.C.Huang,
M.Y.Liu,
W.C.Chang,
and
C.J.Chen
(2007).
Purification, crystallization and preliminary X-ray crystallographic analysis of branched-chain aminotransferase from Deinococcus radiodurans.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
492-494.
|
 |
|
|
|
|
 |
J.A.McCourt,
and
R.G.Duggleby
(2006).
Acetohydroxyacid synthase and its role in the biosynthetic pathway for branched-chain amino acids.
|
| |
Amino Acids,
31,
173-210.
|
 |
|
|
|
|
 |
P.Imming,
C.Sinning,
and
A.Meyer
(2006).
Drugs, their targets and the nature and number of drug targets.
|
| |
Nat Rev Drug Discov,
5,
821-834.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

