 |
PDBsum entry 3daa

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Aminotransferase
|
PDB id
|
|

|

|
3daa
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.6.1.21
- D-amino-acid transaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-alanine + 2-oxoglutarate = D-glutamate + pyruvate
|
 |
 |
 |
 |
 |
 D-alanine
D-alanine
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
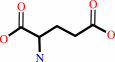 D-glutamate
D-glutamate
|
+
|
 pyruvate
pyruvate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PDD)
matches with 68.18% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
37:4958-4967
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystallographic study of steps along the reaction pathway of D-amino acid aminotransferase.
|
|
D.Peisach,
D.M.Chipman,
P.W.Van Ophem,
J.M.Manning,
D.Ringe.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The three-dimensional structures of two forms of the D-amino acid
aminotransferase (D-aAT) from Bacillus sp. YM-1 have been determined
crystallographically: the pyridoxal phosphate (PLP) form and a complex with the
reduced analogue of the external aldimine, N-(5'-phosphopyridoxyl)-d-alanine
(PPDA). Together with the previously reported pyridoxamine phosphate form of the
enzyme [Sugio et al. (1995) Biochemistry 34, 9661], these structures allow us to
describe the pathway of the enzymatic reaction in structural terms. A major
determinant of the enzyme's stereospecificity for D-amino acids is a group of
three residues (Tyr30, Arg98, and His100, with the latter two contributed by the
neighboring subunit) forming four hydrogen bonds to the substrate alpha-carboxyl
group. The replacement by hydrophobic groups of the homologous residues of the
branched chain L-amino acid aminotransferase (which has a similar fold) could
explain its opposite stereospecificity. As in L-aspartate aminotransferase
(L-AspAT), the cofactor in D-aAT tilts (around its phosphate group and N1 as
pivots) away from the catalytic lysine 145 and the protein face in the course of
the reaction. Unlike L-AspAT, D-aAT shows no other significant conformational
changes during the reaction.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.W.Tremblay,
and
J.S.Blanchard
(2009).
The 1.9 A structure of the branched-chain amino-acid transaminase (IlvE) from Mycobacterium tuberculosis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
1071-1077.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Barreteau,
A.Kovac,
A.Boniface,
M.Sova,
S.Gobec,
and
D.Blanot
(2008).
Cytoplasmic steps of peptidoglycan biosynthesis.
|
| |
FEMS Microbiol Rev,
32,
168-207.
|
 |
|
|
|
|
 |
M.Funakoshi,
M.Sekine,
M.Katane,
T.Furuchi,
M.Yohda,
T.Yoshikawa,
and
H.Homma
(2008).
Cloning and functional characterization of Arabidopsis thaliana D-amino acid aminotransferase--D-aspartate behavior during germination.
|
| |
FEBS J,
275,
1188-1200.
|
 |
|
|
|
|
 |
B.Golinelli-Pimpaneau,
C.Lüthi,
and
P.Christen
(2006).
Structural basis for D-amino acid transamination by the pyridoxal 5'-phosphate-dependent catalytic antibody 15A9.
|
| |
J Biol Chem,
281,
23969-23977.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Goto,
I.Miyahara,
K.Hirotsu,
M.Conway,
N.Yennawar,
M.M.Islam,
and
S.M.Hutson
(2005).
Structural determinants for branched-chain aminotransferase isozyme-specific inhibition by the anticonvulsant drug gabapentin.
|
| |
J Biol Chem,
280,
37246-37256.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.C.Eliot,
and
J.F.Kirsch
(2004).
Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations.
|
| |
Annu Rev Biochem,
73,
383-415.
|
 |
|
|
|
|
 |
M.Noda,
Y.Matoba,
T.Kumagai,
and
M.Sugiyama
(2004).
Structural evidence that alanine racemase from a D-cycloserine-producing microorganism exhibits resistance to its own product.
|
| |
J Biol Chem,
279,
46153-46161.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Kawai,
Y.Ishii,
K.Arakawa,
K.Uemura,
B.Saitoh,
J.Nishimura,
H.Kitazawa,
Y.Yamazaki,
Y.Tateno,
T.Itoh,
and
T.Saito
(2004).
Structural and functional differences in two cyclic bacteriocins with the same sequences produced by lactobacilli.
|
| |
Appl Environ Microbiol,
70,
2906-2911.
|
 |
|
|
|
|
 |
P.Kongsaeree,
C.Samanchart,
P.Laowanapiban,
S.Wiyakrutta,
and
V.Meevootisom
(2003).
Crystallization and preliminary X-ray crystallographic analysis of d-phenylglycine aminotransferase from Pseudomonas stutzeri ST201.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
953-954.
|
 |
|
|
|
|
 |
A.Watanabe,
T.Yoshimura,
B.Mikami,
H.Hayashi,
H.Kagamiyama,
and
N.Esaki
(2002).
Reaction mechanism of alanine racemase from Bacillus stearothermophilus: x-ray crystallographic studies of the enzyme bound with N-(5'-phosphopyridoxyl)alanine.
|
| |
J Biol Chem,
277,
19166-19172.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
O.Hur,
D.Niks,
P.Casino,
and
M.F.Dunn
(2002).
Proton transfers in the beta-reaction catalyzed by tryptophan synthase.
|
| |
Biochemistry,
41,
9991.
|
 |
|
|
|
|
 |
N.Yennawar,
J.Dunbar,
M.Conway,
S.Hutson,
and
G.Farber
(2001).
The structure of human mitochondrial branched-chain aminotransferase.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
506-515.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Christen,
and
P.K.Mehta
(2001).
From cofactor to enzymes. The molecular evolution of pyridoxal-5'-phosphate-dependent enzymes.
|
| |
Chem Rec,
1,
436-447.
|
 |
|
|
|
|
 |
A.Gutierrez,
T.Yoshimura,
Y.Fuchikami,
and
N.Esaki
(2000).
Modulation of activity and substrate specificity by modifying the backbone length of the distant interdomain loop of D-amino acid aminotransferase.
|
| |
Eur J Biochem,
267,
7218-7223.
|
 |
|
|
|
|
 |
K.Kishimoto,
C.Yasuda,
and
J.M.Manning
(2000).
Reversible dissociation/association of D-amino acid transaminase subunits: properties of isolated active dimers and inactive monomers.
|
| |
Biochemistry,
39,
381-387.
|
 |
|
|
|
|
 |
A.A.Morollo,
G.A.Petsko,
and
D.Ringe
(1999).
Structure of a Michaelis complex analogue: propionate binds in the substrate carboxylate site of alanine racemase.
|
| |
Biochemistry,
38,
3293-3301.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.E.Todd,
C.A.Orengo,
and
J.M.Thornton
(1999).
Evolution of protein function, from a structural perspective.
|
| |
Curr Opin Chem Biol,
3,
548-556.
|
 |
|
|
|
|
 |
G.A.Hunter,
and
G.C.Ferreira
(1999).
Pre-steady-state reaction of 5-aminolevulinate synthase. Evidence for a rate-determining product release.
|
| |
J Biol Chem,
274,
12222-12228.
|
 |
|
|
|
|
 |
P.W.van Ophem,
D.Peisach,
S.D.Erickson,
K.Soda,
D.Ringe,
and
J.M.Manning
(1999).
Effects of the E177K mutation in D-amino acid transaminase. Studies on an essential coenzyme anchoring group that contributes to stereochemical fidelity.
|
| |
Biochemistry,
38,
1323-1331.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.N.Jansonius
(1998).
Structure, evolution and action of vitamin B6-dependent enzymes.
|
| |
Curr Opin Struct Biol,
8,
759-769.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

