 |
PDBsum entry 2dki

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2dki
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.13.23
- 3-hydroxybenzoate 4-monooxygenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3-hydroxybenzoate + NADPH + O2 + H+ = 3,4-dihydroxybenzoate + NADP+ + H2O
|
 |
 |
 |
 |
 |
 3-hydroxybenzoate
3-hydroxybenzoate
|
+
|
 NADPH
NADPH
|
+
|
 O2
O2
|
+
|
H(+)
|
=
|
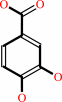 3,4-dihydroxybenzoate
3,4-dihydroxybenzoate
|
+
|
 NADP(+)
NADP(+)
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
364:878-896
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni has a large tunnel for substrate and oxygen access to the active site.
|
|
T.Hiromoto,
S.Fujiwara,
K.Hosokawa,
H.Yamaguchi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The 3-hydroxybenzoate hydroxylase (MHBH) from Comamonas testosteroni KH122-3s is
a single-component flavoprotein monooxygenase, a member of the glutathione
reductase (GR) family. It catalyzes the conversion of 3-hydroxybenzoate to
3,4-dihydroxybenzoate with concomitant requirements for equimolar amounts of
NADPH and molecular oxygen. The production of dihydroxy-benzenoid derivative by
hydroxylation is the first step in the aerobic degradation of various phenolic
compounds in soil microorganisms. To establish the structural basis for
substrate recognition, the crystal structure of MHBH in complex with its
substrate was determined at 1.8 A resolution. The enzyme is shown to form a
physiologically active homodimer with crystallographic 2-fold symmetry, in which
each subunit consists of the first two domains comprising an active site and the
C-terminal domain involved in oligomerization. The protein fold of the catalytic
domains and the active-site architecture, including the FAD and
substrate-binding sites, are similar to those of 4-hydroxybenzoate hydroxylase
(PHBH) and phenol hydroxylase (PHHY), which are members of the GR family,
providing evidence that the flavoprotein aromatic hydroxylases share similar
catalytic actions for hydroxylation of the respective substrates. Structural
comparison of MHBH with the homologous enzymes suggested that a large tunnel
connecting the substrate-binding pocket to the protein surface serves for
substrate transport in this enzyme. The internal space of the large tunnel is
distinctly divided into hydrophilic and hydrophobic regions. The
characteristically stratified environment in the tunnel interior and the size of
the entrance would allow the enzyme to select its substrate by amphiphilic
nature and molecular size. In addition, the structure of the Xe-derivative at
2.5 A resolution led to the identification of a putative oxygen-binding site
adjacent to the substrate-binding pocket. The hydrophobic nature of the
xenon-binding site extends to the solvent through the tunnel, suggesting that
the tunnel could be involved in oxygen transport.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4. Stereo view of the FAD-binding site. The
substrate and FAD molecules are shown as stick models, as in
Figure 2(a). Residues around the FAD-binding site are shown as
stick models and labeled. Water molecules are indicated as red
spheres. The F[o]–F[c] omit electron-density map around the
FAD molecule is colored cyan (countered at the 4.0 σ level).
|
 |
Figure 6.
Figure 6. (a) Superposition of the C^α traces of MHBH and
PHHY (PDB entry 1PN0).^18 MHBH and PHHY are colored blue and
gray, respectively. Each molecule of 3-hydroxybenzoate and FAD
in the MHBH structure is colored purple and yellow,
respectively. For comparison with MHBH, the insertion
segment of PHHY (residues 170–210) is shown in red. (b)
The ribbon diagrams of: left, domain III (residues 453–639) of
MHBH; and right, the human peroxiredoxin, hORF6 (PDB entry
1PRX).^45 Conserved cysteine residues, Cys521 in domain III and
Cys47 located at the active site of hORF6, are shown as stick
models.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2006,
364,
878-896)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.Rosini,
G.Molla,
S.Ghisla,
and
L.Pollegioni
(2011).
On the reaction of D-amino acid oxidase with dioxygen: O2 diffusion pathways and enhancement of reactivity.
|
| |
FEBS J,
278,
482-492.
|
 |
|
|
|
|
 |
G.Volkers,
G.J.Palm,
M.S.Weiss,
G.D.Wright,
and
W.Hinrichs
(2011).
Structural basis for a new tetracycline resistance mechanism relying on the TetX monooxygenase.
|
| |
FEBS Lett,
585,
1061-1066.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Sah,
and
P.S.Phale
(2011).
1-Naphthol 2-hydroxylase from Pseudomonas sp. strain C6: purification, characterization and chemical modification studies.
|
| |
Biodegradation,
22,
517-526.
|
 |
|
|
|
|
 |
M.S.Till,
and
G.M.Ullmann
(2010).
McVol - a program for calculating protein volumes and identifying cavities by a Monte Carlo algorithm.
|
| |
J Mol Model,
16,
419-429.
|
 |
|
|
|
|
 |
U.E.Ukaegbu,
A.Kantz,
M.Beaton,
G.T.Gassner,
and
A.C.Rosenzweig
(2010).
Structure and ligand binding properties of the epoxidase component of styrene monooxygenase .
|
| |
Biochemistry,
49,
1678-1688.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.M.McCulloch,
T.Mukherjee,
T.P.Begley,
and
S.E.Ealick
(2009).
Structure of the PLP degradative enzyme 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti MAFF303099 and its mechanistic implications.
|
| |
Biochemistry,
48,
4139-4149.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.G.Wittmann,
D.Heinrich,
K.Gasow,
A.Frey,
U.Diederichsen,
and
M.G.Rudolph
(2008).
Structures of the human orotidine-5'-monophosphate decarboxylase support a covalent mechanism and provide a framework for drug design.
|
| |
Structure,
16,
82-92.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Chen,
A.Y.Lyubimov,
L.Brammer,
A.Vrielink,
and
N.S.Sampson
(2008).
The binding and release of oxygen and hydrogen peroxide are directed by a hydrophobic tunnel in cholesterol oxidase.
|
| |
Biochemistry,
47,
5368-5377.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Colloc'h,
L.Gabison,
G.Monard,
M.Altarsha,
M.Chiadmi,
G.Marassio,
J.Sopkova-de Oliveira Santos,
M.El Hajji,
B.Castro,
J.H.Abraini,
and
T.Prangé
(2008).
Oxygen pressurized X-ray crystallography: probing the dioxygen binding site in cofactorless urate oxidase and implications for its catalytic mechanism.
|
| |
Biophys J,
95,
2415-2422.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Alfieri,
F.Fersini,
N.Ruangchan,
M.Prongjit,
P.Chaiyen,
and
A.Mattevi
(2007).
Structure of the monooxygenase component of a two-component flavoprotein monooxygenase.
|
| |
Proc Natl Acad Sci U S A,
104,
1177-1182.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.P.Ballou
(2007).
Crystallography gets the jump on the enzymologists.
|
| |
Proc Natl Acad Sci U S A,
104,
15587-15588.
|
 |
|
|
|
|
 |
D.Roeser,
B.Schmidt,
A.Preusser-Kunze,
and
M.G.Rudolph
(2007).
Probing the oxygen-binding site of the human formylglycine-generating enzyme using halide ions.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
621-627.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.S.Ryan,
A.R.Howard-Jones,
M.J.Hamill,
S.J.Elliott,
C.T.Walsh,
and
C.L.Drennan
(2007).
Crystallographic trapping in the rebeccamycin biosynthetic enzyme RebC.
|
| |
Proc Natl Acad Sci U S A,
104,
15311-15316.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.H.Kim,
T.Hisano,
K.Takeda,
W.Iwasaki,
A.Ebihara,
and
K.Miki
(2007).
Crystal structure of the oxygenase component (HpaB) of the 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8.
|
| |
J Biol Chem,
282,
33107-33117.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Y.Kwon,
B.S.Kang,
G.H.Kim,
and
K.J.Kim
(2007).
Expression, purification, crystallization and initial crystallographic characterization of the p-hydroxybenzoate hydroxylase from Corynebacterium glutamicum.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
944-946.
|
 |
|
|
|
|
 |
V.Joosten,
and
W.J.van Berkel
(2007).
Flavoenzymes.
|
| |
Curr Opin Chem Biol,
11,
195-202.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

