 |
PDBsum entry 2qcc

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.4.2.10
- orotate phosphoribosyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pyrimidine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
orotidine 5'-phosphate + diphosphate = orotate + 5-phospho-alpha-D-ribose 1-diphosphate
|
 |
 |
 |
 |
 |
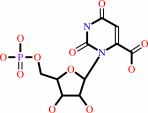 orotidine 5'-phosphate
orotidine 5'-phosphate
|
+
|
 diphosphate
diphosphate
|
=
|
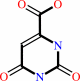 orotate
orotate
|
+
|
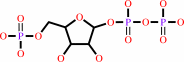 5-phospho-alpha-D-ribose 1-diphosphate
5-phospho-alpha-D-ribose 1-diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.4.1.1.23
- orotidine-5'-phosphate decarboxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
orotidine 5'-phosphate + H+ = UMP + CO2
|
 |
 |
 |
 |
 |
 orotidine 5'-phosphate
orotidine 5'-phosphate
|
+
|
H(+)
|
=
|
 UMP
UMP
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
16:82-92
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structures of the human orotidine-5'-monophosphate decarboxylase support a covalent mechanism and provide a framework for drug design.
|
|
J.G.Wittmann,
D.Heinrich,
K.Gasow,
A.Frey,
U.Diederichsen,
M.G.Rudolph.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
UMP synthase (UMPS) catalyzes the last two steps of de novo pyrimidine
nucleotide synthesis and is a potential cancer drug target. The C-terminal
domain of UMPS is orotidine-5'-monophosphate decarboxylase (OMPD), a
cofactor-less yet extremely efficient enzyme. Studies of OMPDs from
micro-organisms led to the proposal of several noncovalent decarboxylation
mechanisms via high-energy intermediates. We describe nine crystal structures of
human OMPD in complex with substrate, product, and nucleotide inhibitors.
Unexpectedly, simple compounds can replace the natural nucleotides and induce a
closed conformation of OMPD, defining a tripartite catalytic site. The
structures outline the requirements drugs must meet to maximize therapeutic
effects and minimize cross-species activity. Chemical mimicry by iodide
identified a CO(2) product binding site. Plasticity of catalytic residues and a
covalent OMPD-UMP complex prompt a reevaluation of the prevailing
decarboxylation mechanism in favor of covalent intermediates. This mechanism can
also explain the observed catalytic promiscuity of OMPD.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. UMPS Domain Structure and OMPD Biochemistry
(A) Reaction catalyzed by OMPD.
(B) Turnover of OMP
substrate by wild-type OMPD (black) is abolished in the
Asp312Asn mutant (red). If the detection limit of the assay is
assumed to be 5% of the total signal over 4000 s, the Asp312Asn
mutant is at least 1300-fold less active than the wild-type.
(C) Michaelis-Menten kinetics of wild-type OMPD at
25°C.
(D) OMPD is an obligatory dimer of high affinity.
|
 |
Figure 4.
Figure 4. Substrate Binding to OMPD
(A and B) Stereo
representation of the Asp312Asn OMPD-OMP complex showing a bent
carboxylate group.
(C and D) Stereo representation of the
Asp312Asn 6-HMUMP-OMPD complex. The hydroxymethyl group is also
bent out of plane, indicating that electrostatic repulsion by
Asp317b is not responsible for substrate deformation.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Structure
(2008,
16,
82-92)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.J.Wu,
C.C.Liao,
C.H.Jen,
Y.C.Shih,
and
T.C.Chien
(2010).
Chemical models and their mechanistic implications for the transformation of 6-cyanouridine 5'-monophosphate catalyzed by orotidine 5'-monophosphate decarboxylase.
|
| |
Chem Commun (Camb),
46,
4821-4823.
|
 |
|
|
|
|
 |
D.Heinrich,
U.Diederichsen,
and
M.G.Rudolph
(2009).
Lys314 is a nucleophile in non-classical reactions of orotidine-5'-monophosphate decarboxylase.
|
| |
Chemistry,
15,
6619-6625.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Navarrete,
A.Roa,
I.Vaca,
Y.Espinosa,
C.Navarro,
and
R.Chávez
(2009).
Molecular characterization of the niaD and pyrG genes from Penicillium camemberti, and their use as transformation markers.
|
| |
Cell Mol Biol Lett,
14,
692-702.
|
 |
|
|
|
|
 |
Y.Zhang,
M.Luo,
and
V.L.Schramm
(2009).
Transition states of Plasmodium falciparum and human orotate phosphoribosyltransferases.
|
| |
J Am Chem Soc,
131,
4685-4694.
|
 |
|
|
|
|
 |
W.O.Wepukhulu,
V.L.Smiley,
B.Vemulapalli,
J.A.Smiley,
L.M.Phillips,
and
J.K.Lee
(2008).
A substantial oxygen isotope effect at O2 in the OMP decarboxylase reaction: mechanistic implications.
|
| |
Org Biomol Chem,
6,
4533-4541.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

