 |
PDBsum entry 2d3t

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Lyase, oxidoreductase/transferase
|
PDB id
|
|

|

|
2d3t
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase, oxidoreductase/transferase
|
 |
|
Title:
|
 |
Fatty acid beta-oxidation multienzyme complex from pseudomonas fragi, form v
|
|
Structure:
|
 |
Fatty oxidation complex alpha subunit. Chain: a, b. Ec: 4.2.1.17, 5.3.3.8, 1.1.1.35, 5.1.2.3. Engineered: yes. 3-ketoacyl-coa thiolase. Chain: c, d. Synonym: fatty oxidation complex beta subunit, beta-ketothiolase, acetyl-coa acyltransferase. Engineered: yes
|
|
Source:
|
 |
Pseudomonas fragi. Organism_taxid: 296. Expressed in: escherichia coli. Expression_system_taxid: 562. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
Tetramer (from
)
|
|
Resolution:
|
 |
|
3.40Å
|
R-factor:
|
0.242
|
R-free:
|
0.295
|
|
|
Authors:
|
 |
D.Tsuchiya,N.Shimizu,M.Ishikawa,Y.Suzuki,K.Morikawa
|
Key ref:

|
 |
D.Tsuchiya
et al.
(2006).
Ligand-Induced Domain Rearrangement of Fatty Acid beta-Oxidation Multienzyme Complex.
Structure,
14,
237-246.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
01-Oct-05
|
Release date:
|
21-Feb-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chains A, B:
E.C.1.1.1.35
- 3-hydroxyacyl-CoA dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a (3S)-3-hydroxyacyl-CoA + NAD+ = a 3-oxoacyl-CoA + NADH + H+
|
 |
 |
 |
 |
 |
(3S)-3-hydroxyacyl-CoA
Bound ligand (Het Group name = )
matches with 94.44% similarity
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
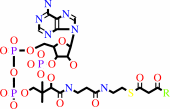 3-oxoacyl-CoA
3-oxoacyl-CoA
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, B:
E.C.4.2.1.17
- enoyl-CoA hydratase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a 4-saturated-(3S)-3-hydroxyacyl-CoA = a (3E)-enoyl-CoA + H2O
|
|
2.
|
a (3S)-3-hydroxyacyl-CoA = a (2E)-enoyl-CoA + H2O
|
|
 |
 |
 |
 |
 |
4-saturated-(3S)-3-hydroxyacyl-CoA
|
=
|
(3E)-enoyl-CoA
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
(3S)-3-hydroxyacyl-CoA
Bound ligand (Het Group name = )
matches with 94.44% similarity
|
=
|
(2E)-enoyl-CoA
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains A, B:
E.C.5.1.2.3
- 3-hydroxybutyryl-CoA epimerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(3S)-3-hydroxybutanoyl-CoA = (3R)-3-hydroxybutanoyl-CoA
|
 |
 |
 |
 |
 |
(3S)-3-hydroxybutanoyl-CoA
|
=
|
(3R)-3-hydroxybutanoyl-CoA
Bound ligand (Het Group name = )
matches with 94.44% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
Chains A, B:
E.C.5.3.3.8
- Delta(3)-Delta(2)-enoyl-CoA isomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a (3Z)-enoyl-CoA = a 4-saturated (2E)-enoyl-CoA
|
|
2.
|
a (3E)-enoyl-CoA = a 4-saturated (2E)-enoyl-CoA
|
|
 |
 |
 |
 |
 |
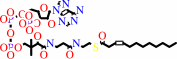 (3Z)-dodec-3-enoyl-CoA
(3Z)-dodec-3-enoyl-CoA
|
=
|
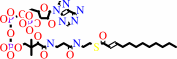 (2E)-dodec-2-enoyl-CoA
(2E)-dodec-2-enoyl-CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
Chains C, D:
E.C.2.3.1.16
- acetyl-CoA C-acyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an acyl-CoA + acetyl-CoA = a 3-oxoacyl-CoA + CoA
|
 |
 |
 |
 |
 |
acyl-CoA
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 acetyl-CoA
acetyl-CoA
|
=
|
 3-oxoacyl-CoA
3-oxoacyl-CoA
|
+
|
 CoA
CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
14:237-246
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Ligand-Induced Domain Rearrangement of Fatty Acid beta-Oxidation Multienzyme Complex.
|
|
D.Tsuchiya,
N.Shimizu,
M.Ishikawa,
Y.Suzuki,
K.Morikawa.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The quaternary structure of a fatty acid beta-oxidation multienzyme complex,
catalyzing three sequential reactions, was investigated by X-ray
crystallographic and small-angle X-ray solution scattering analyses. X-ray
crystallography revealed an intermediate structure of the complex among the
previously reported structures. However, the theoretical scattering curves
calculated from the crystal structures remarkably disagree with the experimental
profiles. Instead, an ensemble of the atomic models, which were all calculated
by rigid-body optimization, reasonably explained the experimental data. These
structures significantly differ from those in the crystals, but they maintain
the substrate binding pocket at the domain boundary. Comparisons among these
structures indicated that binding of 3-hydroxyhexadecanoyl-CoA or nicotinamide
adenine dinucleotide induces domain rearrangements in the complex. The
conformational changes suggest the structural events occurring during the chain
reaction catalyzed by the multienzyme complex.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 2.
Figure 2. Substrate Recognition in the a Subunit
(A)
Local structures around the adenine base binding site in the a
subunit, observed in forms I (red) and V (orange, green). The
bound ligand is acetyl-CoA, which is an analog of
3-hydroxyacyl-CoA.
(B and C) The corresponding structures
of the (B) proximal and (C) distal subunits in form II.
(D)
Superimposition of the two a subunits in the apparently
symmetric form I, in which the acetyl-CoA (the red space-filling
model) was observed. The ligand bound subunit (red) is more open
than the unliganded one (blue). As a consequence, only the aC
domain exhibits a significant difference. The circle with the
broken line indicates the adenine base moiety in (A).
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2006,
14,
237-246)
copyright 2006.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

