 |
PDBsum entry 1yiy

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.6.1.63
- kynurenine--glyoxylate transaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
L-kynurenine + glyoxylate = kynurenate + glycine + H2O
|
|
2.
|
3-hydroxy-L-kynurenine + glyoxylate = xanthurenate + glycine + H2O
|
|
 |
 |
 |
 |
 |
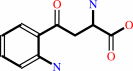 L-kynurenine
L-kynurenine
|
+
|
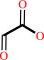 glyoxylate
glyoxylate
|
=
|
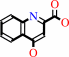 kynurenate
kynurenate
|
+
|
 glycine
glycine
|
+
|
H2O
Bound ligand (Het Group name = )
matches with 57.89% similarity
|
|
 |
 |
 |
 |
 |
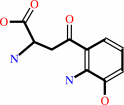 3-hydroxy-L-kynurenine
3-hydroxy-L-kynurenine
|
+
|
 glyoxylate
glyoxylate
|
=
|
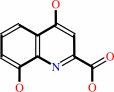 xanthurenate
xanthurenate
|
+
|
 glycine
glycine
|
+
|
H2O
Bound ligand (Het Group name = )
matches with 55.00% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.6.1.7
- kynurenine--oxoglutarate transaminase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-kynurenine + 2-oxoglutarate = kynurenate + L-glutamate + H2O
|
 |
 |
 |
 |
 |
 L-kynurenine
L-kynurenine
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
 kynurenate
kynurenate
|
+
|
 L-glutamate
L-glutamate
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PMP)
matches with 88.24% similarity
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
FEBS J
272:2198-2206
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of Aedes aegypti kynurenine aminotransferase.
|
|
Q.Han,
Y.G.Gao,
H.Robinson,
H.Ding,
S.Wilson,
J.Li.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Aedes aegypti kynurenine aminotransferase (AeKAT) catalyzes the irreversible
transamination of kynurenine to kynurenic acid, the natural antagonist of NMDA
and 7-nicotinic acetycholine receptors. Here, we report the crystal structure of
AeKAT in its PMP and PLP forms at 1.90 and 1.55 A, respectively. The structure
was solved by a combination of single-wavelength anomalous dispersion and
molecular replacement approaches. The initial search model in the molecular
replacement method was built with the result of single-wavelength anomalous
dispersion data from the Br-AeKAT crystal in combination with homology modeling.
The solved structure shows that the enzyme is a homodimer, and that the two
subunits are stabilized by a number of hydrogen bonds, salts bridges, and
hydrophobic interactions. Each subunit is divided into an N-terminal arm and
small and large domains. Based on its folding, the enzyme belongs to the
prototypical fold type, aminotransferase subgroup I. The three-dimensional
structure shows a strictly conserved 'PLP-phosphate binding cup' featuring
PLP-dependent enzymes. The interaction between Cys284 (A) and Cys284 (B) is
unique in AeKAT, which might explain the cysteine effect of AeKAT activity.
Further mutation experiments of this residue are needed to eventually understand
the mechanism of the enzyme modulation by cysteine.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Fig. 3. Diagrams of 2F[o] – F[c] electron density maps
for the active sites of the PMP and PLP forms. The map contoured
at 2.0 sigma is calculated using data between 10.0 and 1.90
Å and 10.0 and 1.55 Å resolution for the PMP and PLP
forms of AeKAT, respectively. (A) PLP form; (B) PMP form.
|
 |
Figure 4.
Fig. 4. Schematic diagram showing active site interactions
in AeKAT. Hydrogen bonds are shown by dotted lines. Phe135 and
Val223 sandwich the pyridine ring of PLP. (A) PLP form; (B) PMP
form.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the Federation of European Biochemical Societies:
FEBS J
(2005,
272,
2198-2206)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Q.Han,
T.Cai,
D.A.Tagle,
and
J.Li
(2010).
Structure, expression, and function of kynurenine aminotransferases in human and rodent brains.
|
| |
Cell Mol Life Sci,
67,
353-368.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Q.Han,
T.Cai,
D.A.Tagle,
and
J.Li
(2010).
Thermal stability, pH dependence and inhibition of four murine kynurenine aminotransferases.
|
| |
BMC Biochem,
11,
19.
|
 |
|
|
|
|
 |
B.Battsetseg,
D.Boldbaatar,
B.Battur,
X.Xuan,
and
K.Fujisaki
(2009).
Cloning and molecular characterization of tick kynurenine aminotransferase (HlKAT) from Haemaphysalis longicornis (Acari: Ixodidae).
|
| |
Parasitol Res,
105,
669-679.
|
 |
|
|
|
|
 |
Q.Han,
H.Robinson,
T.Cai,
D.A.Tagle,
and
J.Li
(2009).
Biochemical and structural properties of mouse kynurenine aminotransferase III.
|
| |
Mol Cell Biol,
29,
784-793.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Q.Han,
H.Robinson,
T.Cai,
D.A.Tagle,
and
J.Li
(2009).
Structural insight into the inhibition of human kynurenine aminotransferase I/glutamine transaminase K.
|
| |
J Med Chem,
52,
2786-2793.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.Rossi,
R.Schwarcz,
and
M.Rizzi
(2008).
Curiosity to kill the KAT (kynurenine aminotransferase): structural insights into brain kynurenic acid synthesis.
|
| |
Curr Opin Struct Biol,
18,
748-755.
|
 |
|
|
|
|
 |
F.Rossi,
S.Garavaglia,
V.Montalbano,
M.A.Walsh,
and
M.Rizzi
(2008).
Crystal Structure of Human Kynurenine Aminotransferase II, a Drug Target for the Treatment of Schizophrenia.
|
| |
J Biol Chem,
283,
3559-3566.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Q.Han,
Y.G.Gao,
H.Robinson,
and
J.Li
(2008).
Structural insight into the mechanism of substrate specificity of aedes kynurenine aminotransferase.
|
| |
Biochemistry,
47,
1622-1630.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Q.Han,
B.T.Beerntsen,
and
J.Li
(2007).
The tryptophan oxidation pathway in mosquitoes with emphasis on xanthurenic acid biosynthesis.
|
| |
J Insect Physiol,
53,
254-263.
|
 |
|
|
|
|
 |
Q.Han,
H.Robinson,
Y.G.Gao,
N.Vogelaar,
S.R.Wilson,
M.Rizzi,
and
J.Li
(2006).
Crystal structures of Aedes aegypti alanine glyoxylate aminotransferase.
|
| |
J Biol Chem,
281,
37175-37182.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

