 |
PDBsum entry 3fvu

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Lyase, transferase
|
PDB id
|
|

|

|
3fvu
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase, transferase
|
 |
|
Title:
|
 |
Crystal structure of human kynurenine aminotransferase i in complex with indole-3-acetic acid
|
|
Structure:
|
 |
Kynurenine--oxoglutarate transaminase 1. Chain: a, b. Synonym: kynurenine--oxoglutarate transaminase i, kynurenine aminotransferase i, kati, glutamine--phenylpyruvate transaminase, glutamine transaminase k, gtk, cysteine-s-conjugate beta-lyase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: ccbl1. Expressed in: spodoptera frugiperda. Expression_system_taxid: 7108.
|
|
Resolution:
|
 |
|
1.55Å
|
R-factor:
|
0.186
|
R-free:
|
0.210
|
|
|
Authors:
|
 |
Q.Han,H.Robinson,T.Cai,D.A.Tagle,J.Li
|
|
Key ref:
|
 |
Q.Han
et al.
(2009).
Structural insight into the inhibition of human kynurenine aminotransferase I/glutamine transaminase K.
J Med Chem,
52,
2786-2793.
PubMed id:
|
 |
|
Date:
|
 |
|
16-Jan-09
|
Release date:
|
19-May-09
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q16773
(KAT1_HUMAN) -
Kynurenine--oxoglutarate transaminase 1 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
422 a.a.
419 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.6.1.64
- glutamine--phenylpyruvate transaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3-phenylpyruvate + L-glutamine = 2-oxoglutaramate + L-phenylalanine
|
 |
 |
 |
 |
 |
3-phenylpyruvate
|
+
|
 L-glutamine
L-glutamine
|
=
|
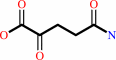 2-oxoglutaramate
2-oxoglutaramate
|
+
|
L-phenylalanine
Bound ligand (Het Group name = )
matches with 78.57% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
Enzyme class 2:
|
 |
E.C.2.6.1.7
- kynurenine--oxoglutarate transaminase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-kynurenine + 2-oxoglutarate = kynurenate + L-glutamate + H2O
|
 |
 |
 |
 |
 |
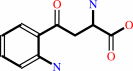 L-kynurenine
L-kynurenine
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
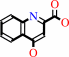 kynurenate
kynurenate
|
+
|
L-glutamate
Bound ligand (Het Group name = )
matches with 68.75% similarity
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
Enzyme class 3:
|
 |
E.C.4.4.1.13
- cysteine-S-conjugate beta-lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an S-substituted L-cysteine + H2O = a thiol + pyruvate + NH4+
|
 |
 |
 |
 |
 |
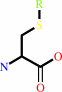 S-substituted L-cysteine
S-substituted L-cysteine
|
+
|
H2O
|
=
|
 thiol
thiol
|
+
|
 pyruvate
pyruvate
|
+
|
NH4(+)
Bound ligand (Het Group name = )
matches with 71.43% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
IAC)
matches with 45.00% similarity
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Med Chem
52:2786-2793
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural insight into the inhibition of human kynurenine aminotransferase I/glutamine transaminase K.
|
|
Q.Han,
H.Robinson,
T.Cai,
D.A.Tagle,
J.Li.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Human kynurenine aminotransferase I (hKAT I) catalyzes the formation of
kynurenic acid, a neuroactive compound. Here, we report three high-resolution
crystal structures (1.50-1.55 A) of hKAT I that are in complex with glycerol and
each of two inhibitors of hKAT I: indole-3-acetic acid (IAC) and Tris. Because
Tris is able to occupy the substrate binding position, we speculate that this
may be the basis for hKAT I inhibition. Furthermore, the hKAT/IAC complex
structure reveals that the binding moieties of the inhibitor are its indole ring
and a carboxyl group. Six chemicals with both binding moieties were tested for
their ability to inhibit hKAT I activity; 3-indolepropionic acid and
DL-indole-3-lactic acid demonstrated the highest level of inhibition, and as
they cannot be considered as substrates of the enzyme, these two inhibitors are
promising candidates for future study. Perhaps even more significantly, we
report the discovery of two different ligands located simultaneously in the hKAT
I active center for the first time.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Q.Han,
H.Robinson,
T.Cai,
D.A.Tagle,
and
J.Li
(2011).
Biochemical and structural characterization of mouse mitochondrial aspartate aminotransferase, a newly identified kynurenine aminotransferase-IV.
|
| |
Biosci Rep,
31,
323-332.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Q.Han,
T.Cai,
D.A.Tagle,
and
J.Li
(2010).
Structure, expression, and function of kynurenine aminotransferases in human and rodent brains.
|
| |
Cell Mol Life Sci,
67,
353-368.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Q.Han,
T.Cai,
D.A.Tagle,
and
J.Li
(2010).
Thermal stability, pH dependence and inhibition of four murine kynurenine aminotransferases.
|
| |
BMC Biochem,
11,
19.
|
 |
|
|
|
|
 |
Z.T.Kincses,
J.Toldi,
and
L.Vécsei
(2010).
Kynurenines, neurodegeneration and Alzheimer's disease.
|
| |
J Cell Mol Med,
14,
2045-2054.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

