 |
PDBsum entry 1vfn

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Nucleoside phosphorylase
|
PDB id
|
|

|

|
1vfn
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.1
- purine-nucleoside phosphorylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a purine D-ribonucleoside + phosphate = a purine nucleobase + alpha- D-ribose 1-phosphate
|
|
2.
|
a purine 2'-deoxy-D-ribonucleoside + phosphate = a purine nucleobase + 2-deoxy-alpha-D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
purine D-ribonucleoside
|
+
|
 phosphate
phosphate
|
=
|
purine nucleobase
|
+
|
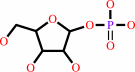 alpha- D-ribose 1-phosphate
alpha- D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
purine 2'-deoxy-D-ribonucleoside
|
+
|
 phosphate
phosphate
|
=
|
purine nucleobase
|
+
|
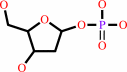 2-deoxy-alpha-D-ribose 1-phosphate
2-deoxy-alpha-D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
265:202-216
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of calf spleen purine nucleoside phosphorylase in a complex with hypoxanthine at 2.15 A resolution.
|
|
G.Koellner,
M.Luić,
D.Shugar,
W.Saenger,
A.Bzowska.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Trimeric calf spleen purine nucleoside phosphorylase has been complexed with
hypoxanthine via phosphorolysis of inosine in the presence of phosphate. The
resulting, "Michaelis" complex (three hypoxanthine molecules per trimer),
presumed to be formed under these conditions, crystallized in the cubic space
group P2(1)3, with unit cell dimension a = 94.11 A and one monomer in the
asymmetric crystal unit; the biologically active trimer is located on the
crystallographic 3-fold axis. High-resolution X-ray diffraction data were
collected using synchrotron radiation (EMBL outstation, Hamburg, c/o DESY). The
crystal structure has been determined by molecular replacement and refined at
2.15 A resolution to an R-value of 0.18. In the hypoxanthine binding site, a
cis-peptide bond between Asn243 and Lys244 is observed. Side-chains of GIu201
and Asn243, as well as one integral water molecule located in the base binding
site, form hydrogen bonds with the hypoxanthine N-1 H, N-7 H and O-6. A second
water molecule links the base positions N-3 and N-9 with an adjacent pocket,
which presumably is the phosphate-binding site. This pocket is filled completely
by a cluster of six water molecules. Hence all possible donor/acceptor-positions
of hypoxanthine are saturated by hydrogen-bonding to protein side-chains or
integral water molecules. Purine nucleoside phosphorylase isolated form human
tissues is a primary target for chemotherapeutic intervention, and the more
stable calf enzyme has similar physico-chemical and kinetic properties, as well
as response to inhibitors. Hence the high-resolution structure presented here
may serve for design of inhibitors with potential pharmacological applications.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4. Mode of binding of the unidentified metal cation
(see the text and Figure 1) located on the 3-fold axis. The
cation is coordinated by His20 N^  epsilon
of three symmetry-related trimers and water molecules Wat429 and
Wat430. Hydrogen bond lengths are denoted in Å (drawn with
SCHAKAL; [Keller 1988]). epsilon
of three symmetry-related trimers and water molecules Wat429 and
Wat430. Hydrogen bond lengths are denoted in Å (drawn with
SCHAKAL; [Keller 1988]).
|
 |
Figure 5.
Figure 5. A drawing of the trimeric calf spleen PNP-Hx
complex. The location of the unidentified metal cation (see also
Figure 2 and Figure 4) is shown as a circle with enlarged van
der Waals radius in each monomer. Direct contacts between
monomers forming the trimer are mainly from the loop between
residues 141 and 168 of one monomer, which is the longest loop
belting the monomer from one side (in red, see also (Table 4 and
Table 4)). Drawn with MOLSCRIPT [Kraulis 1991].
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(1997,
265,
202-216)
copyright 1997.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.F.Visser,
F.Hennessy,
K.Rashamuse,
M.E.Louw,
and
D.Brady
(2010).
Cloning, purification and characterisation of a recombinant purine nucleoside phosphorylase from Bacillus halodurans Alk36.
|
| |
Extremophiles,
14,
185-192.
|
 |
|
|
|
|
 |
H.M.Pereira,
M.M.Rezende,
M.S.Castilho,
G.Oliva,
and
R.C.Garratt
(2010).
Adenosine binding to low-molecular-weight purine nucleoside phosphorylase: the structural basis for recognition based on its complex with the enzyme from Schistosoma mansoni.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
73-79.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Ghanem,
A.S.Murkin,
and
V.L.Schramm
(2009).
Ribocation transition state capture and rebound in human purine nucleoside phosphorylase.
|
| |
Chem Biol,
16,
971-979.
|
 |
|
|
|
|
 |
M.Ghanem,
N.Zhadin,
R.Callender,
and
V.L.Schramm
(2009).
Loop-tryptophan human purine nucleoside phosphorylase reveals submillisecond protein dynamics.
|
| |
Biochemistry,
48,
3658-3668.
|
 |
|
|
|
|
 |
A.Modrak-Wójcik,
A.Kirilenko,
D.Shugar,
and
B.Kierdaszuk
(2008).
Role of ionization of the phosphate cosubstrate on phosphorolysis by purine nucleoside phosphorylase (PNP) of bacterial (E. coli) and mammalian (human) origin.
|
| |
Eur Biophys J,
37,
153-164.
|
 |
|
|
|
|
 |
A.V.Toms,
W.Wang,
Y.Li,
B.Ganem,
and
S.E.Ealick
(2005).
Novel multisubstrate inhibitors of mammalian purine nucleoside phosphorylase.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
1449-1458.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Cacciapuoti,
S.Forte,
M.A.Moretti,
A.Brio,
V.Zappia,
and
M.Porcelli
(2005).
A novel hyperthermostable 5'-deoxy-5'-methylthioadenosine phosphorylase from the archaeon Sulfolobus solfataricus.
|
| |
FEBS J,
272,
1886-1899.
|
 |
|
|
|
|
 |
Y.Zang,
W.H.Wang,
S.W.Wu,
S.E.Ealick,
and
C.C.Wang
(2005).
Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
|
| |
J Biol Chem,
280,
22318-22325.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Luić,
G.Koellner,
T.Yokomatsu,
S.Shibuya,
and
A.Bzowska
(2004).
Calf spleen purine-nucleoside phosphorylase: crystal structure of the binary complex with a potent multisubstrate analogue inhibitor.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1417-1424.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.M.Bennett,
C.Li,
P.W.Allan,
W.B.Parker,
and
S.E.Ealick
(2003).
Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
|
| |
J Biol Chem,
278,
47110-47118.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.M.Pereira,
A.Cleasby,
S.D.Pena S,
G.R.Franco G,
and
R.C.Garratt
(2003).
Cloning, expression and preliminary crystallographic studies of the potential drug target purine nucleoside phosphorylase from Schistosoma mansoni.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
1096-1099.
|
 |
|
|
|
|
 |
G.Stoychev,
B.Kierdaszuk,
and
D.Shugar
(2002).
Xanthosine and xanthine. Substrate properties with purine nucleoside phosphorylases, and relevance to other enzyme systems.
|
| |
Eur J Biochem,
269,
4048-4057.
|
 |
|
|
|
|
 |
K.Lecoq,
I.Belloc,
C.Desgranges,
M.Konrad,
and
B.Daignan-Fornier
(2001).
YLR209c encodes Saccharomyces cerevisiae purine nucleoside phosphorylase.
|
| |
J Bacteriol,
183,
4910-4913.
|
 |
|
|
|
|
 |
M.Luić,
G.Koellner,
D.Shugar,
W.Saenger,
and
A.Bzowska
(2001).
Calf spleen purine nucleoside phosphorylase: structure of its ternary complex with an N(7)-acycloguanosine inhibitor and a phosphate anion.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
30-36.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.C.Appleby,
I.I.Mathews,
M.Porcelli,
G.Cacciapuoti,
and
S.E.Ealick
(2001).
Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
|
| |
J Biol Chem,
276,
39232-39242.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Bzowska,
E.Kulikowska,
and
D.Shugar
(2000).
Purine nucleoside phosphorylases: properties, functions, and clinical aspects.
|
| |
Pharmacol Ther,
88,
349-425.
|
 |
|
|
|
|
 |
F.Wang,
R.W.Miles,
G.Kicska,
E.Nieves,
V.L.Schramm,
and
R.H.Angeletti
(2000).
Immucillin-H binding to purine nucleoside phosphorylase reduces dynamic solvent exchange.
|
| |
Protein Sci,
9,
1660-1668.
|
 |
|
|
|
|
 |
T.C.Appleby,
M.D.Erion,
and
S.E.Ealick
(1999).
The structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase at 1.7 A resolution provides insights into substrate binding and catalysis.
|
| |
Structure,
7,
629-641.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Mao,
W.J.Cook,
M.Zhou,
A.A.Federov,
S.C.Almo,
and
S.E.Ealick
(1998).
Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
|
| |
Biochemistry,
37,
7135-7146.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.S.Hasson,
A.Muscate,
M.J.McLeish,
L.S.Polovnikova,
J.A.Gerlt,
G.L.Kenyon,
G.A.Petsko,
and
D.Ringe
(1998).
The crystal structure of benzoylformate decarboxylase at 1.6 A resolution: diversity of catalytic residues in thiamin diphosphate-dependent enzymes.
|
| |
Biochemistry,
37,
9918-9930.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.W.Miles,
P.C.Tyler,
R.H.Furneaux,
C.K.Bagdassarian,
and
V.L.Schramm
(1998).
One-third-the-sites transition-state inhibitors for purine nucleoside phosphorylase.
|
| |
Biochemistry,
37,
8615-8621.
|
 |
|
|
|
|
 |
Y.Xu,
and
C.Grubmeyer
(1998).
Catalysis in human hypoxanthine-guanine phosphoribosyltransferase: Asp 137 acts as a general acid/base.
|
| |
Biochemistry,
37,
4114-4124.
|
 |
|
|
|
|
 |
C.Mao,
W.J.Cook,
M.Zhou,
G.W.Koszalka,
T.A.Krenitsky,
and
S.E.Ealick
(1997).
The crystal structure of Escherichia coli purine nucleoside phosphorylase: a comparison with the human enzyme reveals a conserved topology.
|
| |
Structure,
5,
1373-1383.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Schmitt,
Y.Mechulam,
M.Fromant,
P.Plateau,
and
S.Blanquet
(1997).
Crystal structure at 1.2 A resolution and active site mapping of Escherichia coli peptidyl-tRNA hydrolase.
|
| |
EMBO J,
16,
4760-4769.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
V.L.Schramm
(1997).
Enzymatic N-riboside scission in RNA and RNA precursors.
|
| |
Curr Opin Chem Biol,
1,
323-331.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

