 |
PDBsum entry 1cb0

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.28
- S-methyl-5'-thioadenosine phosphorylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
S-methyl-5'-thioadenosine + phosphate = 5-(methylsulfanyl)-alpha-D-ribose 1-phosphate + adenine
|
 |
 |
 |
 |
 |
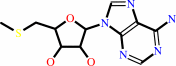 S-methyl-5'-thioadenosine
S-methyl-5'-thioadenosine
|
+
|
 phosphate
phosphate
|
=
|
5-(methylsulfanyl)-alpha-D-ribose 1-phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
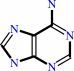 adenine
adenine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
7:629-641
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase at 1.7 A resolution provides insights into substrate binding and catalysis.
|
|
T.C.Appleby,
M.D.Erion,
S.E.Ealick.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: 5'-Deoxy-5'-methylthioadenosine phosphorylase (MTAP) catalyzes the
reversible phosphorolysis of 5'-deoxy-5'-methylthioadenosine (MTA) to adenine
and 5-methylthio-D-ribose-1-phosphate. MTA is a by-product of polyamine
biosynthesis, which is essential for cell growth and proliferation. This salvage
reaction is the principle source of free adenine in human cells. Because of its
importance in coupling the purine salvage pathway to polyamine biosynthesis MTAP
is a potential chemotherapeutic target. RESULTS: We have determined the crystal
structure of MTAP at 1.7 A resolution using multiwavelength anomalous
diffraction phasing techniques. MTAP is a trimer comprised of three identical
subunits. Each subunit consists of a single alpha/beta domain containing a
central eight-stranded mixed beta sheet, a smaller five-stranded mixed beta
sheet and six alpha helices. The native structure revealed the presence of an
adenine molecule in the purine-binding site. The structure of MTAP with
methylthioadenosine and sulfate ion soaked into the active site was also
determined using diffraction data to 1.7 A resolution. CONCLUSIONS: The overall
quaternary structure and subunit topology of MTAP are similar to mammalian
purine nucleoside phosphorylase (PNP). The structures of the MTAP-ligand
complexes provide a map of the active site and suggest possible roles for
specific residues in substrate binding and catalysis. Residues accounting for
the differences in substrate specificity between MTAP and PNP are also
identified. Detailed information about the structure and chemical nature of the
MTAP active site will aid in the rational design of inhibitors of this potential
chemotherapeutic target. The MTAP structure represents the first structure of a
mammalian PNP that is specific for 6-aminopurines.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 3.
Figure 3. Stereoview of the MTAP trimer. The trimer is viewed
down the molecular/crystallographic threefold axis. Each subunit
is shown in a different color, with MTA and sulfate modeled in
red in each of the three active sites. Broken lines indicate
residues 225–229, which are missing in the final model. (The
figure was produced using the program MOLSCRIPT [41].)
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(1999,
7,
629-641)
copyright 1999.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Paul,
S.E.O'Leary,
K.Rajashankar,
W.Bu,
A.Toms,
E.C.Settembre,
J.M.Sanders,
T.P.Begley,
and
S.E.Ealick
(2010).
Glycal formation in crystals of uridine phosphorylase.
|
| |
Biochemistry,
49,
3499-3509.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.R.Ronning,
N.M.Iacopelli,
and
V.Mishra
(2010).
Enzyme-ligand interactions that drive active site rearrangements in the Helicobacter pylori 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
|
| |
Protein Sci,
19,
2498-2510.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Xu
(2010).
Enhancing MAD F(A) data for substructure determination.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
945-949.
|
 |
|
|
|
|
 |
G.Cacciapuoti,
I.Peluso,
F.Fuccio,
and
M.Porcelli
(2009).
Purine nucleoside phosphorylases from hyperthermophilic Archaea require a CXC motif for stability and folding.
|
| |
FEBS J,
276,
5799-5805.
|
 |
|
|
|
|
 |
H.Xu,
and
C.M.Weeks
(2008).
Rapid and automated substructure solution by Shake-and-Bake.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
172-177.
|
 |
|
|
|
|
 |
G.Cacciapuoti,
S.Gorassini,
M.F.Mazzeo,
R.A.Siciliano,
V.Carbone,
V.Zappia,
and
M.Porcelli
(2007).
Biochemical and structural characterization of mammalian-like purine nucleoside phosphorylase from the Archaeon Pyrococcus furiosus.
|
| |
FEBS J,
274,
2482-2495.
|
 |
|
|
|
|
 |
J.L.Jiménez,
B.Hegemann,
J.R.Hutchins,
J.M.Peters,
and
R.Durbin
(2007).
A systematic comparative and structural analysis of protein phosphorylation sites based on the mtcPTM database.
|
| |
Genome Biol,
8,
R90.
|
 |
|
|
|
|
 |
V.Singh,
and
V.L.Schramm
(2006).
Transition-state structure of human 5'-methylthioadenosine phosphorylase.
|
| |
J Am Chem Soc,
128,
14691-14696.
|
 |
|
|
|
|
 |
G.Cacciapuoti,
S.Forte,
M.A.Moretti,
A.Brio,
V.Zappia,
and
M.Porcelli
(2005).
A novel hyperthermostable 5'-deoxy-5'-methylthioadenosine phosphorylase from the archaeon Sulfolobus solfataricus.
|
| |
FEBS J,
272,
1886-1899.
|
 |
|
|
|
|
 |
H.Xu,
C.M.Weeks,
and
H.A.Hauptman
(2005).
Optimizing statistical Shake-and-Bake for Se-atom substructure determination.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
976-981.
|
 |
|
|
|
|
 |
J.E.Lee,
V.Singh,
G.B.Evans,
P.C.Tyler,
R.H.Furneaux,
K.A.Cornell,
M.K.Riscoe,
V.L.Schramm,
and
P.L.Howell
(2005).
Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
|
| |
J Biol Chem,
280,
18274-18282.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.Bu,
E.C.Settembre,
M.H.el Kouni,
and
S.E.Ealick
(2005).
Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
863-872.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Zang,
W.H.Wang,
S.W.Wu,
S.E.Ealick,
and
C.C.Wang
(2005).
Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
|
| |
J Biol Chem,
280,
22318-22325.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Cacciapuoti,
M.A.Moretti,
S.Forte,
A.Brio,
L.Camardella,
V.Zappia,
and
M.Porcelli
(2004).
Methylthioadenosine phosphorylase from the archaeon Pyrococcus furiosus. Mechanism of the reaction and assignment of disulfide bonds.
|
| |
Eur J Biochem,
271,
4834-4844.
|
 |
|
|
|
|
 |
Y.Zhang,
S.E.Cottet,
and
S.E.Ealick
(2004).
Structure of Escherichia coli AMP nucleosidase reveals similarity to nucleoside phosphorylases.
|
| |
Structure,
12,
1383-1394.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.M.Bennett,
C.Li,
P.W.Allan,
W.B.Parker,
and
S.E.Ealick
(2003).
Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
|
| |
J Biol Chem,
278,
47110-47118.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.E.Lee,
K.A.Cornell,
M.K.Riscoe,
and
P.L.Howell
(2003).
Structure of Escherichia coli 5'-methylthioadenosine/ S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis.
|
| |
J Biol Chem,
278,
8761-8770.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Kadariya,
J.Nishioka,
A.Nakamura,
K.Kato-Nakazawa,
and
T.Nobori
(2003).
Molecular characterization of 5'-deoxy-5'-methylthioadenosine phosphorylase-deficient mutant clones of murine lymphoma cell line R1.1.
|
| |
Cancer Sci,
94,
519-522.
|
 |
|
|
|
|
 |
J.Pei,
and
N.V.Grishin
(2002).
Breaking the singleton of germination protease.
|
| |
Protein Sci,
11,
691-697.
|
 |
|
|
|
|
 |
T.C.Appleby,
I.I.Mathews,
M.Porcelli,
G.Cacciapuoti,
and
S.E.Ealick
(2001).
Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
|
| |
J Biol Chem,
276,
39232-39242.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Bzowska,
E.Kulikowska,
and
D.Shugar
(2000).
Purine nucleoside phosphorylases: properties, functions, and clinical aspects.
|
| |
Pharmacol Ther,
88,
349-425.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

