 |
PDBsum entry 1pr5

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.1
- purine-nucleoside phosphorylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a purine D-ribonucleoside + phosphate = a purine nucleobase + alpha- D-ribose 1-phosphate
|
|
2.
|
a purine 2'-deoxy-D-ribonucleoside + phosphate = a purine nucleobase + 2-deoxy-alpha-D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
purine D-ribonucleoside
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
purine nucleobase
|
+
|
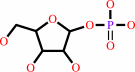 alpha- D-ribose 1-phosphate
alpha- D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
purine 2'-deoxy-D-ribonucleoside
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
purine nucleobase
|
+
|
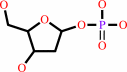 2-deoxy-alpha-D-ribose 1-phosphate
2-deoxy-alpha-D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
278:47110-47118
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
|
|
E.M.Bennett,
C.Li,
P.W.Allan,
W.B.Parker,
S.E.Ealick.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Purine nucleoside phosphorylase catalyzes reversible phosphorolysis of purine
nucleosides and 2'-deoxypurine nucleosides to the free base and ribose (or
2'-deoxyribose) 1-phosphate. Whereas the human enzyme is specific for
6-oxopurine ribonucleosides, the Escherichia coli enzyme accepts additional
substrates including 6-oxopurine ribonucleosides, 6-aminopurine ribonucleosides,
and to a lesser extent purine arabinosides. These differences have been
exploited in a potential suicide gene therapy treatment for solid tumors. In an
effort to optimize this suicide gene therapy approach, we have determined the
three-dimensional structure of the E. coli enzyme in complex with 10 nucleoside
analogs and correlated the structures with kinetic measurements and computer
modeling. These studies explain the preference of the enzyme for ribose sugars,
show increased flexibility for active site residues Asp204 and Arg24, and
suggest that interactions involving the 1- and 6-positions of the purine and the
4'- and 5'-positions of the ribose provide the best opportunities to increase
prodrug specificity and enzyme efficiency.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
FIG. 3. Schematic diagram showing key active site residues
and the contacts that are made with the substrates inosine and
phosphate. Dashed lines indicate hydrogen bonds (or partial
double bonds in carboxylate groups), and W indicates a water
molecule.
|
 |
Figure 7.
FIG. 7. Schematic diagrams of the two possible hydrogen
bonding schemes of the water channel near the purine N-1
position. A, hydrogen bonding scheme in which the first water
molecule is a donor to N-1 of 6-aminopurines (distances are from
the F-Ado A subunit). B, hydrogen bonding scheme in which the
first water molecule is an acceptor from N-1 of 6-oxopurines
(distances are from the Ino C subunit).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2003,
278,
47110-47118)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.P.Nannemann,
K.W.Kaufmann,
J.Meiler,
and
B.O.Bachmann
(2010).
Design and directed evolution of a dideoxy purine nucleoside phosphorylase.
|
| |
Protein Eng Des Sel,
23,
607-616.
|
 |
|
|
|
|
 |
J.Fernández-Lucas,
C.Acebal,
J.V.Sinisterra,
M.Arroyo,
and
I.de la Mata
(2010).
Lactobacillus reuteri 2'-deoxyribosyltransferase, a novel biocatalyst for tailoring of nucleosides.
|
| |
Appl Environ Microbiol,
76,
1462-1470.
|
 |
|
|
|
|
 |
Y.N.Kang,
Y.Zhang,
P.W.Allan,
W.B.Parker,
J.W.Ting,
C.Y.Chang,
and
S.E.Ealick
(2010).
Structure of grouper iridovirus purine nucleoside phosphorylase.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
155-162.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Chaikuad,
and
R.L.Brady
(2009).
Conservation of structure and activity in Plasmodium purine nucleoside phosphorylases.
|
| |
BMC Struct Biol,
9,
42.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Afshar,
M.R.Sawaya,
and
S.L.Morrison
(2009).
Structure of a mutant human purine nucleoside phosphorylase with the prodrug, 2-fluoro-2'-deoxyadenosine and the cytotoxic drug, 2-fluoroadenine.
|
| |
Protein Sci,
18,
1107-1114.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Afshar,
T.Asai,
and
S.L.Morrison
(2009).
Humanized ADEPT comprised of an engineered human purine nucleoside phosphorylase and a tumor targeting peptide for treatment of cancer.
|
| |
Mol Cancer Ther,
8,
185-193.
|
 |
|
|
|
|
 |
A.Modrak-Wójcik,
A.Kirilenko,
D.Shugar,
and
B.Kierdaszuk
(2008).
Role of ionization of the phosphate cosubstrate on phosphorolysis by purine nucleoside phosphorylase (PNP) of bacterial (E. coli) and mammalian (human) origin.
|
| |
Eur Biophys J,
37,
153-164.
|
 |
|
|
|
|
 |
X.Li,
X.Jiang,
H.Li,
and
D.Ren
(2008).
Purine nucleoside phosphorylase from Pseudoalteromonas sp. Bsi590: molecular cloning, gene expression and characterization of the recombinant protein.
|
| |
Extremophiles,
12,
325-333.
|
 |
|
|
|
|
 |
A.Rinaldo-Matthis,
C.Wing,
M.Ghanem,
H.Deng,
P.Wu,
A.Gupta,
P.C.Tyler,
G.B.Evans,
R.H.Furneaux,
S.C.Almo,
C.C.Wang,
and
V.L.Schramm
(2007).
Inhibition and structure of Trichomonas vaginalis purine nucleoside phosphorylase with picomolar transition state analogues.
|
| |
Biochemistry,
46,
659-668.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Toro,
and
E.Grunebaum
(2006).
TAT-mediated intracellular delivery of purine nucleoside phosphorylase corrects its deficiency in mice.
|
| |
J Clin Invest,
116,
2717-2726.
|
 |
|
|
|
|
 |
C.P.Landowski,
X.Song,
P.L.Lorenzi,
J.M.Hilfinger,
and
G.L.Amidon
(2005).
Floxuridine amino acid ester prodrugs: enhancing Caco-2 permeability and resistance to glycosidic bond metabolism.
|
| |
Pharm Res,
22,
1510-1518.
|
 |
|
|
|
|
 |
C.Schnick,
M.A.Robien,
A.M.Brzozowski,
E.J.Dodson,
G.N.Murshudov,
L.Anderson,
J.R.Luft,
C.Mehlin,
W.G.Hol,
J.A.Brannigan,
and
A.J.Wilkinson
(2005).
Structures of Plasmodium falciparum purine nucleoside phosphorylase complexed with sulfate and its natural substrate inosine.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
1245-1254.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Zhang,
S.E.Cottet,
and
S.E.Ealick
(2004).
Structure of Escherichia coli AMP nucleosidase reveals similarity to nucleoside phosphorylases.
|
| |
Structure,
12,
1383-1394.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

