 |
PDBsum entry 1a9o

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Pentosyltransferase
|
PDB id
|
|

|

|
1a9o
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.1
- purine-nucleoside phosphorylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a purine D-ribonucleoside + phosphate = a purine nucleobase + alpha- D-ribose 1-phosphate
|
|
2.
|
a purine 2'-deoxy-D-ribonucleoside + phosphate = a purine nucleobase + 2-deoxy-alpha-D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
purine D-ribonucleoside
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
purine nucleobase
|
+
|
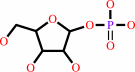 alpha- D-ribose 1-phosphate
alpha- D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
purine 2'-deoxy-D-ribonucleoside
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
purine nucleobase
|
+
|
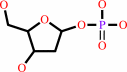 2-deoxy-alpha-D-ribose 1-phosphate
2-deoxy-alpha-D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
37:7135-7146
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
|
|
C.Mao,
W.J.Cook,
M.Zhou,
A.A.Federov,
S.C.Almo,
S.E.Ealick.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Purine nucleoside phosphorylase (PNP) is a key enzyme in the purine salvage
pathway, which provides an alternative to the de novo pathway for the
biosynthesis of purine nucleotides. PNP catalyzes the reversible phosphorolysis
of 2'-deoxypurine ribonucleosides to the free bases and 2-deoxyribose
1-phosphate. Absence of PNP activity in humans is associated with specific
T-cell immune suppression. Its key role in these two processes has made PNP an
important drug design target. We have investigated the structural details of the
PNP-catalyzed reaction by determining the structures of bovine PNP complexes
with various substrates and substrate analogues. The preparation of
phosphate-free crystals of PNP has allowed us to analyze several novel
complexes, including the ternary complex of PNP, purine base, and ribose
1-phosphate and of the completely unbound PNP. These results provide an atomic
view for the catalytic mechanism for PNP proposed by M. D. Erion et al. [(1997)
Biochemistry 36, 11735-11748], in which an oxocarbenium intermediate is
stabilized by phosphate and the negative charge on the purine base is stabilized
by active site residues. The bovine PNP structure reveals several new details of
substrate and inhibitor binding, including two phosphate-induced conformational
changes involving residues 33-36 and 56-69 and a previously undetected role for
His64 in phosphate binding. In addition, a well-ordered water molecule is found
in the PNP active site when purine base or nucleoside is also present. In
contrast to human PNP, only one phosphate binding site was observed. Although
binary complexes were observed for nucleoside, purine base, or phosphate, ribose
1-phosphate binding occurs only in the presence of purine base.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.D'Muniz Pereira,
G.Oliva,
and
R.C.Garratt
(2011).
Purine nucleoside phosphorylase from Schistosoma mansoni in complex with ribose-1-phosphate.
|
| |
J Synchrotron Radiat,
18,
62-65.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.A.Edwards,
J.D.Tipton,
M.D.Brenowitz,
M.R.Emmett,
A.G.Marshall,
G.B.Evans,
P.C.Tyler,
and
V.L.Schramm
(2010).
Conformational states of human purine nucleoside phosphorylase at rest, at work, and with transition state analogues.
|
| |
Biochemistry,
49,
2058-2067.
|
 |
|
|
|
|
 |
D.F.Visser,
F.Hennessy,
K.Rashamuse,
M.E.Louw,
and
D.Brady
(2010).
Cloning, purification and characterisation of a recombinant purine nucleoside phosphorylase from Bacillus halodurans Alk36.
|
| |
Extremophiles,
14,
185-192.
|
 |
|
|
|
|
 |
D.Paul,
S.E.O'Leary,
K.Rajashankar,
W.Bu,
A.Toms,
E.C.Settembre,
J.M.Sanders,
T.P.Begley,
and
S.E.Ealick
(2010).
Glycal formation in crystals of uridine phosphorylase.
|
| |
Biochemistry,
49,
3499-3509.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.N.Kang,
Y.Zhang,
P.W.Allan,
W.B.Parker,
J.W.Ting,
C.Y.Chang,
and
S.E.Ealick
(2010).
Structure of grouper iridovirus purine nucleoside phosphorylase.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
155-162.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.A.Edwards,
J.M.Mason,
K.Clinch,
P.C.Tyler,
G.B.Evans,
and
V.L.Schramm
(2009).
Altered enthalpy-entropy compensation in picomolar transition state analogues of human purine nucleoside phosphorylase.
|
| |
Biochemistry,
48,
5226-5238.
|
 |
|
|
|
|
 |
A.Chaikuad,
and
R.L.Brady
(2009).
Conservation of structure and activity in Plasmodium purine nucleoside phosphorylases.
|
| |
BMC Struct Biol,
9,
42.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Cacciapuoti,
I.Peluso,
F.Fuccio,
and
M.Porcelli
(2009).
Purine nucleoside phosphorylases from hyperthermophilic Archaea require a CXC motif for stability and folding.
|
| |
FEBS J,
276,
5799-5805.
|
 |
|
|
|
|
 |
P.Belenky,
K.C.Christensen,
F.Gazzaniga,
A.A.Pletnev,
and
C.Brenner
(2009).
Nicotinamide riboside and nicotinic acid riboside salvage in fungi and mammals. Quantitative basis for Urh1 and purine nucleoside phosphorylase function in NAD+ metabolism.
|
| |
J Biol Chem,
284,
158-164.
|
 |
|
|
|
|
 |
S.Afshar,
T.Asai,
and
S.L.Morrison
(2009).
Humanized ADEPT comprised of an engineered human purine nucleoside phosphorylase and a tumor targeting peptide for treatment of cancer.
|
| |
Mol Cancer Ther,
8,
185-193.
|
 |
|
|
|
|
 |
A.Modrak-Wójcik,
A.Kirilenko,
D.Shugar,
and
B.Kierdaszuk
(2008).
Role of ionization of the phosphate cosubstrate on phosphorolysis by purine nucleoside phosphorylase (PNP) of bacterial (E. coli) and mammalian (human) origin.
|
| |
Eur Biophys J,
37,
153-164.
|
 |
|
|
|
|
 |
G.Cacciapuoti,
S.Gorassini,
M.F.Mazzeo,
R.A.Siciliano,
V.Carbone,
V.Zappia,
and
M.Porcelli
(2007).
Biochemical and structural characterization of mammalian-like purine nucleoside phosphorylase from the Archaeon Pyrococcus furiosus.
|
| |
FEBS J,
274,
2482-2495.
|
 |
|
|
|
|
 |
J.E.Kim,
and
M.H.Chung
(2006).
8-Oxo-7,8-dihydro-2'-deoxyguanosine is not salvaged for DNA synthesis in human leukemic U937 cells.
|
| |
Free Radic Res,
40,
461-466.
|
 |
|
|
|
|
 |
A.V.Toms,
W.Wang,
Y.Li,
B.Ganem,
and
S.E.Ealick
(2005).
Novel multisubstrate inhibitors of mammalian purine nucleoside phosphorylase.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
1449-1458.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Cacciapuoti,
S.Forte,
M.A.Moretti,
A.Brio,
V.Zappia,
and
M.Porcelli
(2005).
A novel hyperthermostable 5'-deoxy-5'-methylthioadenosine phosphorylase from the archaeon Sulfolobus solfataricus.
|
| |
FEBS J,
272,
1886-1899.
|
 |
|
|
|
|
 |
W.Bu,
E.C.Settembre,
M.H.el Kouni,
and
S.E.Ealick
(2005).
Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
863-872.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Cacciapuoti,
M.A.Moretti,
S.Forte,
A.Brio,
L.Camardella,
V.Zappia,
and
M.Porcelli
(2004).
Methylthioadenosine phosphorylase from the archaeon Pyrococcus furiosus. Mechanism of the reaction and assignment of disulfide bonds.
|
| |
Eur J Biochem,
271,
4834-4844.
|
 |
|
|
|
|
 |
M.Luić,
G.Koellner,
T.Yokomatsu,
S.Shibuya,
and
A.Bzowska
(2004).
Calf spleen purine-nucleoside phosphorylase: crystal structure of the binary complex with a potent multisubstrate analogue inhibitor.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1417-1424.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Lewandowicz,
P.C.Tyler,
G.B.Evans,
R.H.Furneaux,
and
V.L.Schramm
(2003).
Achieving the ultimate physiological goal in transition state analogue inhibitors for purine nucleoside phosphorylase.
|
| |
J Biol Chem,
278,
31465-31468.
|
 |
|
|
|
|
 |
E.M.Bennett,
C.Li,
P.W.Allan,
W.B.Parker,
and
S.E.Ealick
(2003).
Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
|
| |
J Biol Chem,
278,
47110-47118.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Stoychev,
B.Kierdaszuk,
and
D.Shugar
(2002).
Xanthosine and xanthine. Substrate properties with purine nucleoside phosphorylases, and relevance to other enzyme systems.
|
| |
Eur J Biochem,
269,
4048-4057.
|
 |
|
|
|
|
 |
A.D.Andricopulo,
and
R.A.Yunes
(2001).
Structure-activity relationships for a collection of structurally diverse inhibitors of purine nucleoside phosphorylase.
|
| |
Chem Pharm Bull (Tokyo),
49,
10-17.
|
 |
|
|
|
|
 |
A.Fedorov,
W.Shi,
G.Kicska,
E.Fedorov,
P.C.Tyler,
R.H.Furneaux,
J.C.Hanson,
G.J.Gainsford,
J.Z.Larese,
V.L.Schramm,
and
S.C.Almo
(2001).
Transition state structure of purine nucleoside phosphorylase and principles of atomic motion in enzymatic catalysis.
|
| |
Biochemistry,
40,
853-860.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Lecoq,
I.Belloc,
C.Desgranges,
M.Konrad,
and
B.Daignan-Fornier
(2001).
YLR209c encodes Saccharomyces cerevisiae purine nucleoside phosphorylase.
|
| |
J Bacteriol,
183,
4910-4913.
|
 |
|
|
|
|
 |
M.Luić,
G.Koellner,
D.Shugar,
W.Saenger,
and
A.Bzowska
(2001).
Calf spleen purine nucleoside phosphorylase: structure of its ternary complex with an N(7)-acycloguanosine inhibitor and a phosphate anion.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
30-36.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.C.Appleby,
I.I.Mathews,
M.Porcelli,
G.Cacciapuoti,
and
S.E.Ealick
(2001).
Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
|
| |
J Biol Chem,
276,
39232-39242.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Bzowska,
E.Kulikowska,
and
D.Shugar
(2000).
Purine nucleoside phosphorylases: properties, functions, and clinical aspects.
|
| |
Pharmacol Ther,
88,
349-425.
|
 |
|
|
|
|
 |
F.Wang,
R.W.Miles,
G.Kicska,
E.Nieves,
V.L.Schramm,
and
R.H.Angeletti
(2000).
Immucillin-H binding to purine nucleoside phosphorylase reduces dynamic solvent exchange.
|
| |
Protein Sci,
9,
1660-1668.
|
 |
|
|
|
|
 |
M.J.Pugmire,
and
S.E.Ealick
(1998).
The crystal structure of pyrimidine nucleoside phosphorylase in a closed conformation.
|
| |
Structure,
6,
1467-1479.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

