 |
PDBsum entry 1bfd

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.1.7
- benzoylformate decarboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
phenylglyoxylate + H+ = benzaldehyde + CO2
|
 |
 |
 |
 |
 |
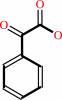 phenylglyoxylate
phenylglyoxylate
|
+
|
H(+)
|
=
|
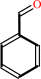 benzaldehyde
benzaldehyde
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Thiamine diphosphate
|
 |
 |
 |
 |
 |
Thiamine diphosphate
Bound ligand (Het Group name =
TPP)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
37:9918-9930
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
The crystal structure of benzoylformate decarboxylase at 1.6 A resolution: diversity of catalytic residues in thiamin diphosphate-dependent enzymes.
|
|
M.S.Hasson,
A.Muscate,
M.J.McLeish,
L.S.Polovnikova,
J.A.Gerlt,
G.L.Kenyon,
G.A.Petsko,
D.Ringe.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of the thiamin diphosphate (ThDP)-dependent enzyme
benzoylformate decarboxylase (BFD), the third enzyme in the mandelate pathway of
Pseudomonas putida, has been solved by multiple isomorphous replacement at 1.6 A
resolution and refined to an R-factor of 15.0% (free R = 18.6%). The structure
of BFD has been compared to that of other ThDP-dependent enzymes, including
pyruvate decarboxylase. The overall architecture of BFD resembles that of the
other family members, and cofactor- and metal-binding residues are well
conserved. Surprisingly, there is no conservation of active-site residues not
directly bound to the cofactor. The position of functional groups in the active
site may be conserved, however. Three classes of metal ions have been identified
in the BFD crystal structure: Ca2+ bound to the cofactor in each subunit, Mg2+
on a 2-fold axis of the tetramer, and Ca2+ at a crystal contact. The structure
includes a non-proline cis-peptide bond and an unusually long and regular
polyproline type II helix that mediates the main contact between tetramers in
the crystal. The high-quality electron-density map allowed the correction of
errors totaling more than 10% of the amino acid sequence, which had been
predicted from the reported sequence of the mdlC gene. Analysis of the BFD
structure suggests that requirements for activation of the cofactor, the nature
of the reaction intermediates, and architectural considerations relating to the
protein fold have been dominant forces in the evolution of ThDP-dependent
enzymes.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.G.Topal,
C.Atilgan,
A.S.Demir,
and
V.Aviyente
(2010).
Understanding the mode of action of ThDP in benzoylformate decarboxylase.
|
| |
Biopolymers,
93,
32-46.
|
 |
|
|
|
|
 |
T.G.Palmen,
J.Nieveler,
B.Frölich,
W.Treffenfeldt,
M.Pohl,
and
J.Büchs
(2010).
Physiological relation between respiration activity and heterologous expression of selected benzoylformate decarboxylase variants in Escherichia coli.
|
| |
Microb Cell Fact,
9,
76.
|
 |
|
|
|
|
 |
X.Y.Pei,
K.M.Erixon,
B.F.Luisi,
and
F.J.Leeper
(2010).
Structural insights into the prereaction state of pyruvate decarboxylase from Zymomonas mobilis .
|
| |
Biochemistry,
49,
1727-1736.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.T.Lefort,
Y.M.Hyun,
J.B.Schultz,
F.Y.Law,
R.E.Waugh,
P.A.Knauf,
and
M.Kim
(2009).
Outside-in signal transmission by conformational changes in integrin Mac-1.
|
| |
J Immunol,
183,
6460-6468.
|
 |
|
|
|
|
 |
G.S.Brandt,
M.M.Kneen,
S.Chakraborty,
A.T.Baykal,
N.Nemeria,
A.Yep,
D.I.Ruby,
G.A.Petsko,
G.L.Kenyon,
M.J.McLeish,
F.Jordan,
and
D.Ringe
(2009).
Snapshot of a reaction intermediate: analysis of benzoylformate decarboxylase in complex with a benzoylphosphonate inhibitor.
|
| |
Biochemistry,
48,
3247-3257.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Müller,
D.Gocke,
and
M.Pohl
(2009).
Thiamin diphosphate in biological chemistry: exploitation of diverse thiamin diphosphate-dependent enzymes for asymmetric chemoenzymatic synthesis.
|
| |
FEBS J,
276,
2894-2904.
|
 |
|
|
|
|
 |
R.J.Mikolajek,
A.C.Spiess,
M.Pohl,
and
J.Büchs
(2009).
Propioin synthesis using thiamine diphosphate-dependent enzymes.
|
| |
Biotechnol Prog,
25,
132-138.
|
 |
|
|
|
|
 |
S.Chakraborty,
N.S.Nemeria,
A.Balakrishnan,
G.S.Brandt,
M.M.Kneen,
A.Yep,
M.J.McLeish,
G.L.Kenyon,
G.A.Petsko,
D.Ringe,
and
F.Jordan
(2009).
Detection and time course of formation of major thiamin diphosphate-bound covalent intermediates derived from a chromophoric substrate analogue on benzoylformate decarboxylase.
|
| |
Biochemistry,
48,
981-994.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.M.Hyun,
H.L.Chung,
J.L.McGrath,
R.E.Waugh,
and
M.Kim
(2009).
Activated integrin VLA-4 localizes to the lamellipodia and mediates T cell migration on VCAM-1.
|
| |
J Immunol,
183,
359-369.
|
 |
|
|
|
|
 |
A.Yep,
G.L.Kenyon,
and
M.J.McLeish
(2008).
Saturation mutagenesis of putative catalytic residues of benzoylformate decarboxylase provides a challenge to the accepted mechanism.
|
| |
Proc Natl Acad Sci U S A,
105,
5733-5738.
|
 |
|
|
|
|
 |
D.Gocke,
L.Walter,
E.Gauchenova,
G.Kolter,
M.Knoll,
C.L.Berthold,
G.Schneider,
J.Pleiss,
M.Müller,
and
M.Pohl
(2008).
Rational protein design of ThDP-dependent enzymes-engineering stereoselectivity.
|
| |
Chembiochem,
9,
406-412.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Kluger,
and
S.Rathgeber
(2008).
Catalyzing separation of carbon dioxide in thiamin diphosphate-promoted decarboxylation.
|
| |
FEBS J,
275,
6089-6100.
|
 |
|
|
|
|
 |
S.C.Wenzel,
H.B.Bode,
I.Kochems,
and
R.Müller
(2008).
A type I/type III polyketide synthase hybrid biosynthetic pathway for the structurally unique ansa compound kendomycin.
|
| |
Chembiochem,
9,
2711-2721.
|
 |
|
|
|
|
 |
S.J.Costelloe,
J.M.Ward,
and
P.A.Dalby
(2008).
Evolutionary Analysis of the TPP-Dependent Enzyme Family.
|
| |
J Mol Evol,
66,
36-49.
|
 |
|
|
|
|
 |
V.I.Bunik,
and
D.Degtyarev
(2008).
Structure-function relationships in the 2-oxo acid dehydrogenase family: substrate-specific signatures and functional predictions for the 2-oxoglutarate dehydrogenase-like proteins.
|
| |
Proteins,
71,
874-890.
|
 |
|
|
|
|
 |
C.L.Berthold,
D.Gocke,
M.D.Wood,
F.J.Leeper,
M.Pohl,
and
G.Schneider
(2007).
Structure of the branched-chain keto acid decarboxylase (KdcA) from Lactococcus lactis provides insights into the structural basis for the chemoselective and enantioselective carboligation reaction.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
1217-1224.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.C.Juan,
M.M.Hoque,
M.T.Hossain,
T.Yamamoto,
S.Imamura,
K.Suzuki,
T.Sekiguchi,
and
A.Takénaka
(2007).
The structures of pyruvate oxidase from Aerococcus viridans with cofactors and with a reaction intermediate reveal the flexibility of the active-site tunnel for catalysis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
900-907.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Xie,
S.Vucetic,
L.M.Iakoucheva,
C.J.Oldfield,
A.K.Dunker,
Z.Obradovic,
and
V.N.Uversky
(2007).
Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins.
|
| |
J Proteome Res,
6,
1917-1932.
|
 |
|
|
|
|
 |
R.Mikolajek,
A.C.Spiess,
M.Pohl,
S.Lamare,
and
J.Büchs
(2007).
An activity, stability and selectivity comparison of propioin synthesis by thiamine diphosphate-dependent enzymes in a solid/gas bioreactor.
|
| |
Chembiochem,
8,
1063-1070.
|
 |
|
|
|
|
 |
S.Spaepen,
W.Versées,
D.Gocke,
M.Pohl,
J.Steyaert,
and
J.Vanderleyden
(2007).
Characterization of phenylpyruvate decarboxylase, involved in auxin production of Azospirillum brasilense.
|
| |
J Bacteriol,
189,
7626-7633.
|
 |
|
|
|
|
 |
S.Watanabe,
R.Matsumi,
T.Arai,
H.Atomi,
T.Imanaka,
and
K.Miki
(2007).
Crystal structures of [NiFe] hydrogenase maturation proteins HypC, HypD, and HypE: insights into cyanation reaction by thiol redox signaling.
|
| |
Mol Cell,
27,
29-40.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.Versées,
S.Spaepen,
J.Vanderleyden,
and
J.Steyaert
(2007).
The crystal structure of phenylpyruvate decarboxylase from Azospirillum brasilense at 1.5 A resolution. Implications for its catalytic and regulatory mechanism.
|
| |
FEBS J,
274,
2363-2375.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Malandrinos,
M.Louloudi,
and
N.Hadjiliadis
(2006).
Thiamine models and perspectives on the mechanism of action of thiamine-dependent enzymes.
|
| |
Chem Soc Rev,
35,
684-692.
|
 |
|
|
|
|
 |
H.Henning,
C.Leggewie,
M.Pohl,
M.Müller,
T.Eggert,
and
K.E.Jaeger
(2006).
Identification of novel benzoylformate decarboxylases by growth selection.
|
| |
Appl Environ Microbiol,
72,
7510-7517.
|
 |
|
|
|
|
 |
J.A.McCourt,
and
R.G.Duggleby
(2006).
Acetohydroxyacid synthase and its role in the biosynthetic pathway for branched-chain amino acids.
|
| |
Amino Acids,
31,
173-210.
|
 |
|
|
|
|
 |
J.A.McCourt,
S.S.Pang,
J.King-Scott,
L.W.Guddat,
and
R.G.Duggleby
(2006).
Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase.
|
| |
Proc Natl Acad Sci U S A,
103,
569-573.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Goto,
H.Hayashi,
I.Miyahara,
K.Hirotsu,
M.Yoshida,
and
T.Oikawa
(2006).
Crystal structures of nonoxidative zinc-dependent 2,6-dihydroxybenzoate (gamma-resorcylate) decarboxylase from Rhizobium sp. strain MTP-10005.
|
| |
J Biol Chem,
281,
34365-34373.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Knoll,
M.Müller,
J.Pleiss,
and
M.Pohl
(2006).
Factors mediating activity, selectivity, and substrate specificity for the thiamin diphosphate-dependent enzymes benzaldehyde lyase and benzoylformate decarboxylase.
|
| |
Chembiochem,
7,
1928-1934.
|
 |
|
|
|
|
 |
C.L.Berthold,
P.Moussatche,
N.G.Richards,
and
Y.Lindqvist
(2005).
Structural basis for activation of the thiamin diphosphate-dependent enzyme oxalyl-CoA decarboxylase by adenosine diphosphate.
|
| |
J Biol Chem,
280,
41645-41654.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.V.Cubellis,
F.Caillez,
T.L.Blundell,
and
S.C.Lovell
(2005).
Properties of polyproline II, a secondary structure element implicated in protein-protein interactions.
|
| |
Proteins,
58,
880-892.
|
 |
|
|
|
|
 |
N.Nemeria,
K.Tittmann,
E.Joseph,
L.Zhou,
M.B.Vazquez-Coll,
P.Arjunan,
G.Hübner,
W.Furey,
and
F.Jordan
(2005).
Glutamate 636 of the Escherichia coli pyruvate dehydrogenase-E1 participates in active center communication and behaves as an engineered acetolactate synthase with unusual stereoselectivity.
|
| |
J Biol Chem,
280,
21473-21482.
|
 |
|
|
|
|
 |
R.Golbik,
L.E.Meshalkina,
T.Sandalova,
K.Tittmann,
E.Fiedler,
H.Neef,
S.König,
R.Kluger,
G.A.Kochetov,
G.Schneider,
and
G.Hübner
(2005).
Effect of coenzyme modification on the structural and catalytic properties of wild-type transketolase and of the variant E418A from Saccharomyces cerevisiae.
|
| |
FEBS J,
272,
1326-1342.
|
 |
|
|
|
|
 |
T.G.Mosbacher,
M.Mueller,
and
G.E.Schulz
(2005).
Structure and mechanism of the ThDP-dependent benzaldehyde lyase from Pseudomonas fluorescens.
|
| |
FEBS J,
272,
6067-6076.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.E.Caines,
J.M.Elkins,
K.S.Hewitson,
and
C.J.Schofield
(2004).
Crystal structure and mechanistic implications of N2-(2-carboxyethyl)arginine synthase, the first enzyme in the clavulanic acid biosynthesis pathway.
|
| |
J Biol Chem,
279,
5685-5692.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.S.Pang,
R.G.Duggleby,
R.L.Schowen,
and
L.W.Guddat
(2004).
The crystal structures of Klebsiella pneumoniae acetolactate synthase with enzyme-bound cofactor and with an unusual intermediate.
|
| |
J Biol Chem,
279,
2242-2253.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.M.Ciszak,
L.G.Korotchkina,
P.M.Dominiak,
S.Sidhu,
and
M.S.Patel
(2003).
Structural basis for flip-flop action of thiamin pyrophosphate-dependent enzymes revealed by human pyruvate dehydrogenase.
|
| |
J Biol Chem,
278,
21240-21246.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Zhang,
J.Dai,
Z.Lu,
and
D.Dunaway-Mariano
(2003).
The phosphonopyruvate decarboxylase from Bacteroides fragilis.
|
| |
J Biol Chem,
278,
41302-41308.
|
 |
|
|
|
|
 |
M.Bhasin,
J.L.Billinsky,
and
D.R.Palmer
(2003).
Steady-state kinetics and molecular evolution of Escherichia coli MenD [(1R,6R)-2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase], an anomalous thiamin diphosphate-dependent decarboxylase-carboligase.
|
| |
Biochemistry,
42,
13496-13504.
|
 |
|
|
|
|
 |
M.Fries,
H.J.Chauhan,
G.J.Domingo,
H.I.Jung,
and
R.N.Perham
(2003).
Site-directed mutagenesis of a loop at the active site of E1 (alpha2beta2) of the pyruvate dehydrogenase complex. A possible common sequence motif.
|
| |
Eur J Biochem,
270,
861-870.
|
 |
|
|
|
|
 |
M.J.McLeish,
M.M.Kneen,
K.N.Gopalakrishna,
C.W.Koo,
P.C.Babbitt,
J.A.Gerlt,
and
G.L.Kenyon
(2003).
Identification and characterization of a mandelamide hydrolase and an NAD(P)+-dependent benzaldehyde dehydrogenase from Pseudomonas putida ATCC 12633.
|
| |
J Bacteriol,
185,
2451-2456.
|
 |
|
|
|
|
 |
S.S.Pang,
L.W.Guddat,
and
R.G.Duggleby
(2003).
Molecular basis of sulfonylurea herbicide inhibition of acetohydroxyacid synthase.
|
| |
J Biol Chem,
278,
7639-7644.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Y.Huang,
A.K.Chang,
P.F.Nixon,
and
R.G.Duggleby
(2001).
Site-directed mutagenesis of the ionizable groups in the active site of Zymomonas mobilis pyruvate decarboxylase: effect on activity and pH dependence.
|
| |
Eur J Biochem,
268,
3558-3565.
|
 |
|
|
|
|
 |
J.Wang,
R.Golbik,
B.Seliger,
M.Spinka,
K.Tittmann,
G.Hübner,
and
F.Jordan
(2001).
Consequences of a modified putative substrate-activation site on catalysis by yeast pyruvate decarboxylase.
|
| |
Biochemistry,
40,
1755-1763.
|
 |
|
|
|
|
 |
L.J.Baker,
J.A.Dorocke,
R.A.Harris,
and
D.E.Timm
(2001).
The crystal structure of yeast thiamin pyrophosphokinase.
|
| |
Structure,
9,
539-546.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Nemeria,
Y.Yan,
Z.Zhang,
A.M.Brown,
P.Arjunan,
W.Furey,
J.R.Guest,
and
F.Jordan
(2001).
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by thiamin 2-thiazolone and thiamin 2-thiothiazolone diphosphates. Evidence for reversible tight-binding inhibition.
|
| |
J Biol Chem,
276,
45969-45978.
|
 |
|
|
|
|
 |
S.S.Pang,
L.W.Guddat,
and
R.G.Duggleby
(2001).
Crystallization of the catalytic subunit of Saccharomyces cerevisiae acetohydroxyacid synthase.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
1321-1323.
|
 |
|
|
|
|
 |
A.AEvarsson,
J.L.Chuang,
R.M.Wynn,
S.Turley,
D.T.Chuang,
and
W.G.Hol
(2000).
Crystal structure of human branched-chain alpha-ketoacid dehydrogenase and the molecular basis of multienzyme complex deficiency in maple syrup urine disease.
|
| |
Structure,
8,
277-291.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.A.Sergienko,
J.Wang,
L.Polovnikova,
M.S.Hasson,
M.J.McLeish,
G.L.Kenyon,
and
F.Jordan
(2000).
Spectroscopic detection of transient thiamin diphosphate-bound intermediates on benzoylformate decarboxylase.
|
| |
Biochemistry,
39,
13862-13869.
|
 |
|
|
|
|
 |
O.P.Ward,
and
A.Singh
(2000).
Enzymatic asymmetric synthesis by decarboxylases.
|
| |
Curr Opin Biotechnol,
11,
520-526.
|
 |
|
|
|
|
 |
A.E.Todd,
C.A.Orengo,
and
J.M.Thornton
(1999).
Evolution of protein function, from a structural perspective.
|
| |
Curr Opin Chem Biol,
3,
548-556.
|
 |
|
|
|
|
 |
C.A.Orengo,
A.E.Todd,
and
J.M.Thornton
(1999).
From protein structure to function.
|
| |
Curr Opin Struct Biol,
9,
374-382.
|
 |
|
|
|
|
 |
M.H.Charon,
A.Volbeda,
E.Chabriere,
L.Pieulle,
and
J.C.Fontecilla-Camps
(1999).
Structure and electron transfer mechanism of pyruvate:ferredoxin oxidoreductase.
|
| |
Curr Opin Struct Biol,
9,
663-669.
|
 |
|
|
|
|
 |
G.Schenk,
R.G.Duggleby,
and
P.F.Nixon
(1998).
Properties and functions of the thiamin diphosphate dependent enzyme transketolase.
|
| |
Int J Biochem Cell Biol,
30,
1297-1318.
|
 |
|
|
|
|
 |
M.S.Hasson,
I.Schlichting,
J.Moulai,
K.Taylor,
W.Barrett,
G.L.Kenyon,
P.C.Babbitt,
J.A.Gerlt,
G.A.Petsko,
and
D.Ringe
(1998).
Evolution of an enzyme active site: the structure of a new crystal form of muconate lactonizing enzyme compared with mandelate racemase and enolase.
|
| |
Proc Natl Acad Sci U S A,
95,
10396-10401.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

