 |
PDBsum entry 1od2

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.6.3.4.14
- biotin carboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N6-biotinyl-L-lysyl-[protein] + hydrogencarbonate + ATP = N6- carboxybiotinyl-L-lysyl-[protein] + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
N(6)-biotinyl-L-lysyl-[protein]
|
+
|
 hydrogencarbonate
hydrogencarbonate
|
+
|
 ATP
ATP
|
=
|
N(6)- carboxybiotinyl-L-lysyl-[protein]
Bound ligand (Het Group name = )
matches with 56.25% similarity
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.6.4.1.2
- acetyl-CoA carboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
hydrogencarbonate + acetyl-CoA + ATP = malonyl-CoA + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
hydrogencarbonate
Bound ligand (Het Group name = )
matches with 94.12% similarity
|
+
|
 acetyl-CoA
acetyl-CoA
|
+
|
 ATP
ATP
|
=
|
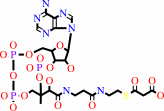 malonyl-CoA
malonyl-CoA
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Biotin
|
 |
 |
 |
 |
 |
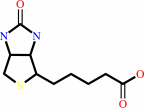 Biotin
Biotin
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Science
299:2064-2067
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase.
|
|
H.Zhang,
Z.Yang,
Y.Shen,
L.Tong.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Acetyl-coenzyme A carboxylases (ACCs) are required for the biosynthesis and
oxidation of long-chain fatty acids. They are targets for therapeutics against
obesity and diabetes, and several herbicides function by inhibiting their
carboxyltransferase (CT) domain. We determined the crystal structure of the free
enzyme and the coenzyme A complex of yeast CT at 2.7 angstrom resolution and
found that it comprises two domains, both belonging to the crotonase/ClpP
superfamily. The active site is at the interface of a dimer. Mutagenesis and
kinetic studies reveal the functional roles of conserved residues here. The
herbicides target the active site of CT, providing a lead for inhibitor
development against human ACCs.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Structures of ACCs. (A) Schematic drawing of the
primary structures of eukaryotic multidomain ACC and bacterial
multisubunit ACC. (B) The chemical reaction catalyzed by CT. The
N1 atom of biotin is labeled. (C) Schematic drawing of the
structure of the CT domain dimer of yeast ACC. The N and C
domains of one monomer are colored cyan and yellow, whereas
those of the other monomer are colored purple and green. The CoA
molecule bound to one monomer is shown as a stick model. Only
the adenine base was observed in the other monomer (labeled A).
(C) was produced with Ribbons (22).
|
 |
Figure 3.
Fig. 3. The active site of CT and the binding mode of CoA. (A)
Schematic drawing in stereo of the active site of CT. The N
domain is shown in cyan, and the C domain of the other monomer
is shown in green. The side chains of residues in the active
site are shown in purple. The prime (') in the labels indicates
the C domain of the other monomer of the dimer. (B) Molecular
surface of the active site region of CT. The side chain of
Lys1764 (in helix  6, 15
Å from the active site) has been removed to facilitate the
viewing of the active site. (C) Chemical structure of haloxyfop
and the double reciprocal plot showing the competitive
inhibition of wild-type yeast CT by haloxyfop. (A) was produced
with Ribbons (22), and (B) with Grasp (23). 6, 15
Å from the active site) has been removed to facilitate the
viewing of the active site. (C) Chemical structure of haloxyfop
and the double reciprocal plot showing the competitive
inhibition of wild-type yeast CT by haloxyfop. (A) was produced
with Ribbons (22), and (B) with Grasp (23).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the AAAs:
Science
(2003,
299,
2064-2067)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.F.Franca,
F.L.Leite,
R.A.Cunha,
O.N.Oliveira,
and
L.C.Freitas
(2011).
Designing an enzyme-based nanobiosensor using molecular modeling techniques.
|
| |
Phys Chem Chem Phys,
13,
8894-8899.
|
 |
|
|
|
|
 |
G.Gago,
L.Diacovich,
A.Arabolaza,
S.C.Tsai,
and
H.Gramajo
(2011).
Fatty acid biosynthesis in actinomycetes.
|
| |
FEMS Microbiol Rev,
35,
475-497.
|
 |
|
|
|
|
 |
C.L.Colbert,
C.W.Kim,
Y.A.Moon,
L.Henry,
M.Palnitkar,
W.B.McKean,
K.Fitzgerald,
J.Deisenhofer,
J.D.Horton,
and
H.J.Kwon
(2010).
Crystal structure of Spot 14, a modulator of fatty acid synthesis.
|
| |
Proc Natl Acad Sci U S A,
107,
18820-18825.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.S.Huang,
K.Sadre-Bazzaz,
Y.Shen,
B.Deng,
Z.H.Zhou,
and
L.Tong
(2010).
Crystal structure of the alpha(6)beta(6) holoenzyme of propionyl-coenzyme A carboxylase.
|
| |
Nature,
466,
1001-1005.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.P.Yu,
Y.S.Kim,
and
L.Tong
(2010).
Mechanism for the inhibition of the carboxyltransferase domain of acetyl-coenzyme A carboxylase by pinoxaden.
|
| |
Proc Natl Acad Sci U S A,
107,
22072-22077.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.B.Powles,
and
Q.Yu
(2010).
Evolution in action: plants resistant to herbicides.
|
| |
Annu Rev Plant Biol,
61,
317-347.
|
 |
|
|
|
|
 |
C.Y.Chou,
L.P.Yu,
and
L.Tong
(2009).
Crystal structure of biotin carboxylase in complex with substrates and implications for its catalytic mechanism.
|
| |
J Biol Chem,
284,
11690-11697.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Kress,
D.Brügel,
I.Schall,
D.Linder,
W.Buckel,
and
L.O.Essen
(2009).
An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases.
|
| |
J Biol Chem,
284,
28401-28409.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Bains,
R.Leon,
and
M.J.Boulanger
(2009).
Structural and biophysical characterization of BoxC from Burkholderia xenovorans LB400: a novel ring-cleaving enzyme in the crotonase superfamily.
|
| |
J Biol Chem,
284,
16377-16385.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.C.Castle,
Y.Hara,
C.K.Raymond,
P.Garrett-Engele,
K.Ohwaki,
Z.Kan,
J.Kusunoki,
and
J.M.Johnson
(2009).
ACC2 is expressed at high levels human white adipose and has an isoform with a novel N-terminus.
|
| |
PLoS ONE,
4,
e4369.
|
 |
|
|
|
|
 |
K.P.Madauss,
W.A.Burkhart,
T.G.Consler,
D.J.Cowan,
W.K.Gottschalk,
A.B.Miller,
S.A.Short,
T.B.Tran,
and
S.P.Williams
(2009).
The human ACC2 CT-domain C-terminus is required for full functionality and has a novel twist.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
449-461.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Xiang,
M.M.Callaghan,
K.G.Watson,
and
L.Tong
(2009).
A different mechanism for the inhibition of the carboxyltransferase domain of acetyl-coenzyme A carboxylase by tepraloxydim.
|
| |
Proc Natl Acad Sci U S A,
106,
20723-20727.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.K.Benson,
G.Meades,
A.Grove,
and
G.L.Waldrop
(2008).
DNA inhibits catalysis by the carboxyltransferase subunit of acetyl-CoA carboxylase: implications for active site communication.
|
| |
Protein Sci,
17,
34-42.
|
 |
|
|
|
|
 |
X.Liu,
P.D.Fortin,
and
C.T.Walsh
(2008).
Andrimid producers encode an acetyl-CoA carboxyltransferase subunit resistant to the action of the antibiotic.
|
| |
Proc Natl Acad Sci U S A,
105,
13321-13326.
|
 |
|
|
|
|
 |
A.A.Hoskins,
M.Morar,
T.J.Kappock,
I.I.Mathews,
J.B.Zaugg,
T.E.Barder,
P.Peng,
A.Okamoto,
S.E.Ealick,
and
J.Stubbe
(2007).
N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis.
|
| |
Biochemistry,
46,
2842-2855.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Yamada,
R.Natsume,
T.Nakamatsu,
S.Horinouchi,
H.Kawasaki,
and
T.Senda
(2007).
Crystallization and preliminary crystallographic analysis of DtsR1, a carboxyltransferase subunit of acetyl-CoA carboxylase from Corynebacterium glutamicum.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
120-122.
|
 |
|
|
|
|
 |
L.Tong,
and
H.J.Harwood
(2006).
Acetyl-coenzyme A carboxylases: versatile targets for drug discovery.
|
| |
J Cell Biochem,
99,
1476-1488.
|
 |
|
|
|
|
 |
T.W.Lin,
M.M.Melgar,
D.Kurth,
S.J.Swamidass,
J.Purdon,
T.Tseng,
G.Gago,
P.Baldi,
H.Gramajo,
and
S.C.Tsai
(2006).
Structure-based inhibitor design of AccD5, an essential acyl-CoA carboxylase carboxyltransferase domain of Mycobacterium tuberculosis.
|
| |
Proc Natl Acad Sci U S A,
103,
3072-3077.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Shen,
C.Y.Chou,
G.G.Chang,
and
L.Tong
(2006).
Is dimerization required for the catalytic activity of bacterial biotin carboxylase?
|
| |
Mol Cell,
22,
807-818.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Zhang,
B.Tweel,
J.Li,
and
L.Tong
(2004).
Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase in complex with CP-640186.
|
| |
Structure,
12,
1683-1691.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Zhang,
B.Tweel,
and
L.Tong
(2004).
Molecular basis for the inhibition of the carboxyltransferase domain of acetyl-coenzyme-A carboxylase by haloxyfop and diclofop.
|
| |
Proc Natl Acad Sci U S A,
101,
5910-5915.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Kondo,
Y.Nakajima,
S.Sugio,
J.Yong-Biao,
S.Sueda,
and
H.Kondo
(2004).
Structure of the biotin carboxylase subunit of pyruvate carboxylase from Aquifex aeolicus at 2.2 A resolution.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
486-492.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Kanamori,
N.Kanou,
H.Atomi,
and
T.Imanaka
(2004).
Enzymatic characterization of a prokaryotic urea carboxylase.
|
| |
J Bacteriol,
186,
2532-2539.
|
 |
|
|
|
|
 |
Y.Sasaki,
and
Y.Nagano
(2004).
Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding.
|
| |
Biosci Biotechnol Biochem,
68,
1175-1184.
|
 |
|
|
|
|
 |
Y.Shen,
S.L.Volrath,
S.C.Weatherly,
T.D.Elich,
and
L.Tong
(2004).
A mechanism for the potent inhibition of eukaryotic acetyl-coenzyme A carboxylase by soraphen A, a macrocyclic polyketide natural product.
|
| |
Mol Cell,
16,
881-891.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Shi,
and
P.Burn
(2004).
Lipid metabolic enzymes: emerging drug targets for the treatment of obesity.
|
| |
Nat Rev Drug Discov,
3,
695-710.
|
 |
|
|
|
|
 |
K.S.Wendt,
I.Schall,
R.Huber,
W.Buckel,
and
U.Jacob
(2003).
Crystal structure of the carboxyltransferase subunit of the bacterial sodium ion pump glutaconyl-coenzyme A decarboxylase.
|
| |
EMBO J,
22,
3493-3502.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

